Genetically engineered microorganisms for the detection of explosives' residues
- PMID: 26579085
- PMCID: PMC4625088
- DOI: 10.3389/fmicb.2015.01175
Genetically engineered microorganisms for the detection of explosives' residues
Abstract
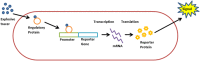
The manufacture and use of explosives throughout the past century has resulted in the extensive pollution of soils and groundwater, and the widespread interment of landmines imposes a major humanitarian risk and prevents civil development of large areas. As most current landmine detection technologies require actual presence at the surveyed areas, thus posing a significant risk to personnel, diverse research efforts are aimed at the development of remote detection solutions. One possible means proposed to fulfill this objective is the use of microbial bioreporters: genetically engineered microorganisms "tailored" to generate an optical signal in the presence of explosives' vapors. The use of such sensor bacteria will allow to pinpoint the locations of explosive devices in a minefield. While no study has yet resulted in a commercially operational system, significant progress has been made in the design and construction of explosives-sensing bacterial strains. In this article we review the attempts to construct microbial bioreporters for the detection of explosives, and analyze the steps that need to be undertaken for this strategy to be applicable for landmine detection.
Keywords: 2,4,6- trinitrotoluene; 2,4-dinitrotoluene; bioluminescence; biosensors; explosives; landmines; microbial bioreporters.
Figures
Similar articles
-
Bacterial bioreporters for the detection of trace explosives: performance enhancement by DNA shuffling and random mutagenesis.Appl Microbiol Biotechnol. 2021 May;105(10):4329-4337. doi: 10.1007/s00253-021-11290-2. Epub 2021 May 4. Appl Microbiol Biotechnol. 2021. PMID: 33942130
-
Detection of buried explosives with immobilized bacterial bioreporters.Microb Biotechnol. 2021 Jan;14(1):251-261. doi: 10.1111/1751-7915.13683. Epub 2020 Oct 23. Microb Biotechnol. 2021. PMID: 33095504 Free PMC article.
-
Microbial bioreporters of trace explosives.Curr Opin Biotechnol. 2017 Jun;45:113-119. doi: 10.1016/j.copbio.2017.03.003. Epub 2017 Mar 17. Curr Opin Biotechnol. 2017. PMID: 28319855 Review.
-
Introduction of quorum sensing elements into bacterial bioreporter circuits enhances explosives' detection capabilities.Eng Life Sci. 2022 Mar 2;22(3-4):308-318. doi: 10.1002/elsc.202100134. eCollection 2022 Mar. Eng Life Sci. 2022. PMID: 35382532 Free PMC article.
-
Controlled biological and biomimetic systems for landmine detection.Biosens Bioelectron. 2007 Aug 30;23(1):1-18. doi: 10.1016/j.bios.2007.05.005. Epub 2007 Jun 2. Biosens Bioelectron. 2007. PMID: 17662594 Review.
Cited by
-
Enhancing DNT Detection by a Bacterial Bioreporter: Directed Evolution of the Transcriptional Activator YhaJ.Front Bioeng Biotechnol. 2022 Feb 14;10:821835. doi: 10.3389/fbioe.2022.821835. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35237579 Free PMC article.
-
Synthetic Biology Enables Programmable Cell-Based Biosensors.Chemphyschem. 2020 Jan 16;21(2):132-144. doi: 10.1002/cphc.201900739. Epub 2019 Oct 25. Chemphyschem. 2020. PMID: 31585026 Free PMC article. Review.
-
Bacterial bioreporters for the detection of trace explosives: performance enhancement by DNA shuffling and random mutagenesis.Appl Microbiol Biotechnol. 2021 May;105(10):4329-4337. doi: 10.1007/s00253-021-11290-2. Epub 2021 May 4. Appl Microbiol Biotechnol. 2021. PMID: 33942130
-
Detection of buried explosives with immobilized bacterial bioreporters.Microb Biotechnol. 2021 Jan;14(1):251-261. doi: 10.1111/1751-7915.13683. Epub 2020 Oct 23. Microb Biotechnol. 2021. PMID: 33095504 Free PMC article.
-
A Portable Biosensor for 2,4-Dinitrotoluene Vapors.Sensors (Basel). 2018 Dec 3;18(12):4247. doi: 10.3390/s18124247. Sensors (Basel). 2018. PMID: 30513956 Free PMC article.
References
-
- Altamirano M., García-Villada L., Agrelo M., Sánchez-Martín L., Martín-Otero L., Flores-Moya A., et al. (2004). A novel approach to improve specificity of algal biosensors using wild-type and resistant mutants: an application to detect TNT. Biosens. Bioelectron. 19 1319–1323. 10.1016/j.bios.2003.4460.11.001 - DOI - PubMed
-
- Belkin S., Smulski D. R., Dadon S., Vollmer A. C., Van Dyk T. K., Larossa R. A. (1997). A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Water Res. 31 3009–3016. 10.1016/S0043-1354(97)00169-3 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

