Distribution of circular proteins in plants: large-scale mapping of cyclotides in the Violaceae
- PMID: 26579135
- PMCID: PMC4621522
- DOI: 10.3389/fpls.2015.00855
Distribution of circular proteins in plants: large-scale mapping of cyclotides in the Violaceae
Abstract
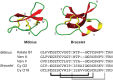
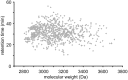
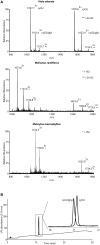
During the last decade there has been increasing interest in small circular proteins found in plants of the violet family (Violaceae). These so-called cyclotides consist of a circular chain of approximately 30 amino acids, including six cysteines forming three disulfide bonds, arranged in a cyclic cystine knot (CCK) motif. In this study we map the occurrence and distribution of cyclotides throughout the Violaceae. Plant material was obtained from herbarium sheets containing samples up to 200 years of age. Even the oldest specimens contained cyclotides in the preserved leaves, with no degradation products observable, confirming their place as one of the most stable proteins in nature. Over 200 samples covering 17 of the 23-31 genera in Violaceae were analyzed, and cyclotides were positively identified in 150 species. Each species contained a unique set of between one and 25 cyclotides, with many exclusive to individual plant species. We estimate the number of different cyclotides in the Violaceae to be 5000-25,000, and propose that cyclotides are ubiquitous among all Violaceae species. Twelve new cyclotides from six phylogenetically dispersed genera were sequenced. Furthermore, the first glycosylated derivatives of cyclotides were identified and characterized, further increasing the diversity and complexity of this unique protein family.
Keywords: Violaceae; cyclotide; plant peptide; plant protein; stable proteins.
Figures


Similar articles
-
Cyclotide proteins and precursors from the genus Gloeospermum: filling a blank spot in the cyclotide map of Violaceae.Phytochemistry. 2010 Jan;71(1):13-20. doi: 10.1016/j.phytochem.2009.09.023. Epub 2009 Oct 30. Phytochemistry. 2010. PMID: 19879608
-
Circular proteins from Melicytus (Violaceae) refine the conserved protein and gene architecture of cyclotides.Org Biomol Chem. 2009 Jun 7;7(11):2378-88. doi: 10.1039/b823020j. Epub 2009 Apr 6. Org Biomol Chem. 2009. PMID: 19462049
-
Understanding the Diversity and Distribution of Cyclotides from Plants of Varied Genetic Origin.J Nat Prod. 2017 May 26;80(5):1522-1530. doi: 10.1021/acs.jnatprod.7b00061. Epub 2017 May 4. J Nat Prod. 2017. PMID: 28471681
-
Novel strategies for isolation and characterization of cyclotides: the discovery of bioactive macrocyclic plant polypeptides in the Violaceae.Curr Protein Pept Sci. 2004 Oct;5(5):317-29. doi: 10.2174/1389203043379495. Curr Protein Pept Sci. 2004. PMID: 15544528 Review.
-
Discovery, structure, function, and applications of cyclotides: circular proteins from plants.J Exp Bot. 2016 Aug;67(16):4801-12. doi: 10.1093/jxb/erw210. Epub 2016 May 23. J Exp Bot. 2016. PMID: 27222514 Review.
Cited by
-
The involvement of cyclotides in mutual interactions of violets and the two-spotted spider mite.Sci Rep. 2022 Feb 3;12(1):1914. doi: 10.1038/s41598-022-05461-y. Sci Rep. 2022. PMID: 35115562 Free PMC article.
-
Gas-Phase Sequencing of Cyclotides: Introduction of Selective Ring Opening at Dehydroalanine via Ion/Ion Reaction.Anal Chem. 2019 Dec 17;91(24):15608-15616. doi: 10.1021/acs.analchem.9b03671. Epub 2019 Dec 3. Anal Chem. 2019. PMID: 31746593 Free PMC article.
-
Transcriptomics Identifies Modules of Differentially Expressed Genes and Novel Cyclotides in Viola pubescens.Front Plant Sci. 2019 Feb 15;10:156. doi: 10.3389/fpls.2019.00156. eCollection 2019. Front Plant Sci. 2019. PMID: 30828342 Free PMC article.
-
In vitro Inhibition of HIV-1 by Cyclotide-Enriched Extracts of Viola tricolor.Front Pharmacol. 2022 May 27;13:888961. doi: 10.3389/fphar.2022.888961. eCollection 2022. Front Pharmacol. 2022. PMID: 35712712 Free PMC article.
-
Molecular Chimera in Cancer Drug Discovery: Beyond Antibody Therapy, Designing Grafted Stable Peptides Targeting Cancer.Int J Pept Res Ther. 2025;31(3):38. doi: 10.1007/s10989-025-10690-6. Epub 2025 Feb 17. Int J Pept Res Ther. 2025. PMID: 39974747 Free PMC article. Review.
References
-
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., et al. . (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. 10.1039/C2NP20085F - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

