Transcriptional regulation of drought response: a tortuous network of transcriptional factors
- PMID: 26579147
- PMCID: PMC4625044
- DOI: 10.3389/fpls.2015.00895
Transcriptional regulation of drought response: a tortuous network of transcriptional factors
Abstract
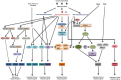
Drought is one of the leading factors responsible for the reduction in crop yield worldwide. Due to climate change, in future, more areas are going to be affected by drought and for prolonged periods. Therefore, understanding the mechanisms underlying the drought response is one of the major scientific concerns for improving crop yield. Plants deploy diverse strategies and mechanisms to respond and tolerate drought stress. Expression of numerous genes is modulated in different plants under drought stress that help them to optimize their growth and development. Plant hormone abscisic acid (ABA) plays a major role in plant response and tolerance by regulating the expression of many genes under drought stress. Transcription factors being the major regulator of gene expression play a crucial role in stress response. ABA regulates the expression of most of the target genes through ABA-responsive element (ABRE) binding protein/ABRE binding factor (AREB/ABF) transcription factors. Genes regulated by AREB/ABFs constitute a regulon termed as AREB/ABF regulon. In addition to this, drought responsive genes are also regulated by ABA-independent mechanisms. In ABA-independent regulation, dehydration-responsive element binding protein (DREB), NAM, ATAF, and CUC regulons play an important role by regulating many drought-responsive genes. Apart from these major regulons, MYB/MYC, WRKY, and nuclear factor-Y (NF-Y) transcription factors are also involved in drought response and tolerance. Our understanding about transcriptional regulation of drought is still evolving. Recent reports have suggested the existence of crosstalk between different transcription factors operating under drought stress. In this article, we have reviewed various regulons working under drought stress and their crosstalk with each other.
Keywords: ABA; cross-talk; drought; regulons; transcription factors.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

