Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes
- PMID: 26579161
- PMCID: PMC4626571
- DOI: 10.3389/fpls.2015.00914
Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes
Abstract
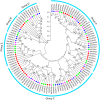
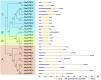
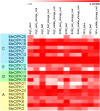
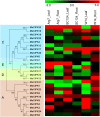
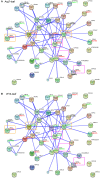
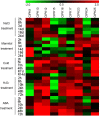
Cassava is an important food and potential biofuel crop that is tolerant to multiple abiotic stressors. The mechanisms underlying these tolerances are currently less known. CBL-interacting protein kinases (CIPKs) have been shown to play crucial roles in plant developmental processes, hormone signaling transduction, and in the response to abiotic stress. However, no data is currently available about the CPK family in cassava. In this study, a total of 25 CIPK genes were identified from cassava genome based on our previous genome sequencing data. Phylogenetic analysis suggested that 25 MeCIPKs could be classified into four subfamilies, which was supported by exon-intron organizations and the architectures of conserved protein motifs. Transcriptomic analysis of a wild subspecies and two cultivated varieties showed that most MeCIPKs had different expression patterns between wild subspecies and cultivatars in different tissues or in response to drought stress. Some orthologous genes involved in CIPK interaction networks were identified between Arabidopsis and cassava. The interaction networks and co-expression patterns of these orthologous genes revealed that the crucial pathways controlled by CIPK networks may be involved in the differential response to drought stress in different accessions of cassava. Nine MeCIPK genes were selected to investigate their transcriptional response to various stimuli and the results showed the comprehensive response of the tested MeCIPK genes to osmotic, salt, cold, oxidative stressors, and ABA signaling. The identification and expression analysis of CIPK family suggested that CIPK genes are important components of development and multiple signal transduction pathways in cassava. The findings of this study will help lay a foundation for the functional characterization of the CIPK gene family and provide an improved understanding of abiotic stress responses and signaling transduction in cassava.
Keywords: CBL-interacting protein kinases; abiotic stress; cassava; gene expression; genome-wide; identification.
Figures

Similar articles
-
Genome-Wide Identification and Expression Analysis of the Mitogen-Activated Protein Kinase Gene Family in Cassava.Front Plant Sci. 2016 Aug 30;7:1294. doi: 10.3389/fpls.2016.01294. eCollection 2016. Front Plant Sci. 2016. PMID: 27625666 Free PMC article.
-
Expression Patterns and Identified Protein-Protein Interactions Suggest That Cassava CBL-CIPK Signal Networks Function in Responses to Abiotic Stresses.Front Plant Sci. 2018 Mar 2;9:269. doi: 10.3389/fpls.2018.00269. eCollection 2018. Front Plant Sci. 2018. PMID: 29552024 Free PMC article.
-
Genome-wide survey and expression analysis of the calcium-dependent protein kinase gene family in cassava.Mol Genet Genomics. 2016 Feb;291(1):241-53. doi: 10.1007/s00438-015-1103-x. Epub 2015 Aug 14. Mol Genet Genomics. 2016. PMID: 26272723
-
Molecular Mechanisms of CBL-CIPK Signaling Pathway in Plant Abiotic Stress Tolerance and Hormone Crosstalk.Int J Mol Sci. 2024 May 6;25(9):5043. doi: 10.3390/ijms25095043. Int J Mol Sci. 2024. PMID: 38732261 Free PMC article. Review.
-
The CBL-CIPK Pathway in Plant Response to Stress Signals.Int J Mol Sci. 2020 Aug 7;21(16):5668. doi: 10.3390/ijms21165668. Int J Mol Sci. 2020. PMID: 32784662 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Expression Analysis of the KUP Family under Abiotic Stress in Cassava (Manihot esculenta Crantz).Front Physiol. 2018 Jan 24;9:17. doi: 10.3389/fphys.2018.00017. eCollection 2018. Front Physiol. 2018. PMID: 29416511 Free PMC article.
-
Comprehensive transcriptional and functional analyses of melatonin synthesis genes in cassava reveal their novel role in hypersensitive-like cell death.Sci Rep. 2016 Oct 14;6:35029. doi: 10.1038/srep35029. Sci Rep. 2016. PMID: 27739451 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the WRKY Gene Family in Cassava.Front Plant Sci. 2016 Feb 5;7:25. doi: 10.3389/fpls.2016.00025. eCollection 2016. Front Plant Sci. 2016. PMID: 26904033 Free PMC article.
-
Identification, expression, and association analysis of calcineurin B-like protein-interacting protein kinase genes in peanut.Front Genet. 2022 Sep 5;13:939255. doi: 10.3389/fgene.2022.939255. eCollection 2022. Front Genet. 2022. PMID: 36134030 Free PMC article.
-
MeCIPK23 interacts with Whirly transcription factors to activate abscisic acid biosynthesis and regulate drought resistance in cassava.Plant Biotechnol J. 2020 Jul;18(7):1504-1506. doi: 10.1111/pbi.13321. Epub 2020 Jan 19. Plant Biotechnol J. 2020. PMID: 31858710 Free PMC article. No abstract available.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

