Remodeling of nuclear landscapes during human myelopoietic cell differentiation maintains co-aligned active and inactive nuclear compartments
- PMID: 26579212
- PMCID: PMC4647504
- DOI: 10.1186/s13072-015-0038-0
Remodeling of nuclear landscapes during human myelopoietic cell differentiation maintains co-aligned active and inactive nuclear compartments
Abstract
Background: Previous studies of higher order chromatin organization in nuclei of mammalian species revealed both structural consistency and species-specific differences between cell lines and during early embryonic development. Here, we extended our studies to nuclear landscapes in the human myelopoietic lineage representing a somatic cell differentiation system. Our longterm goal is a search for structural features of nuclei, which are restricted to certain cell types/species, as compared to features, which are evolutionary highly conserved, arguing for their basic functional roles in nuclear organization.
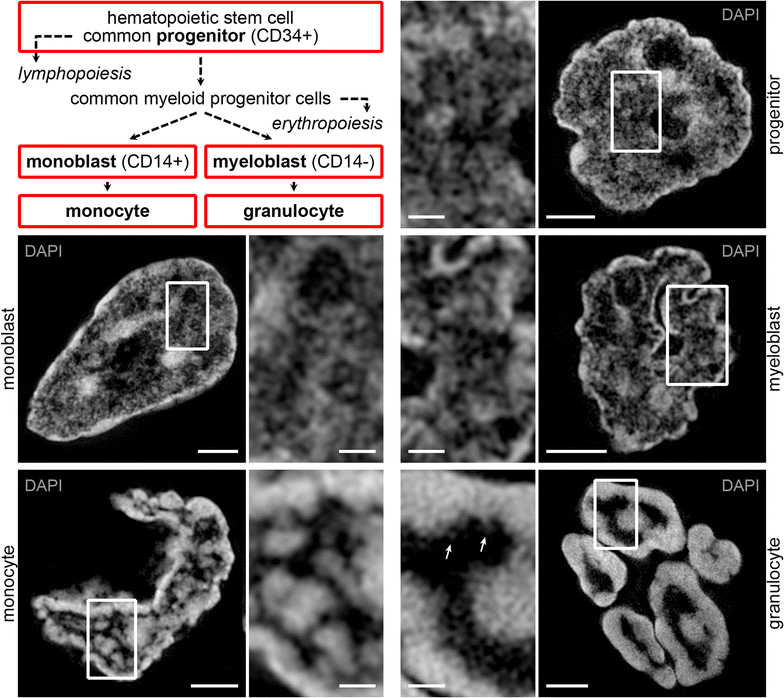
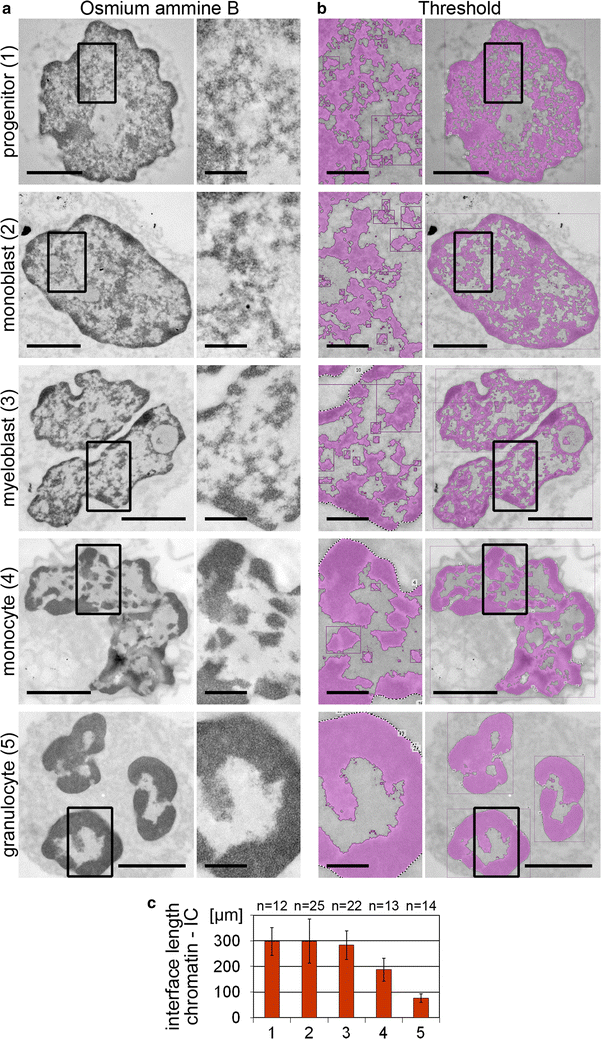
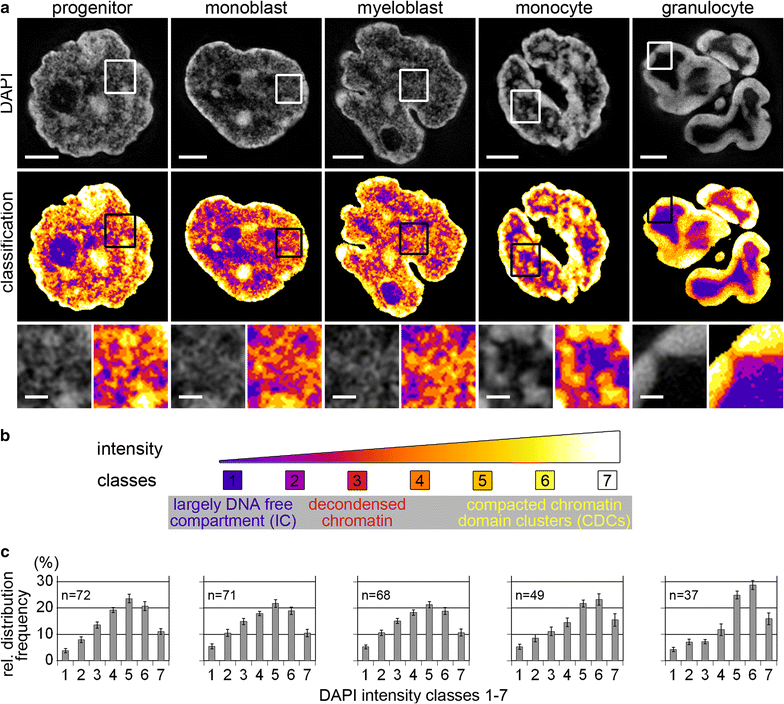
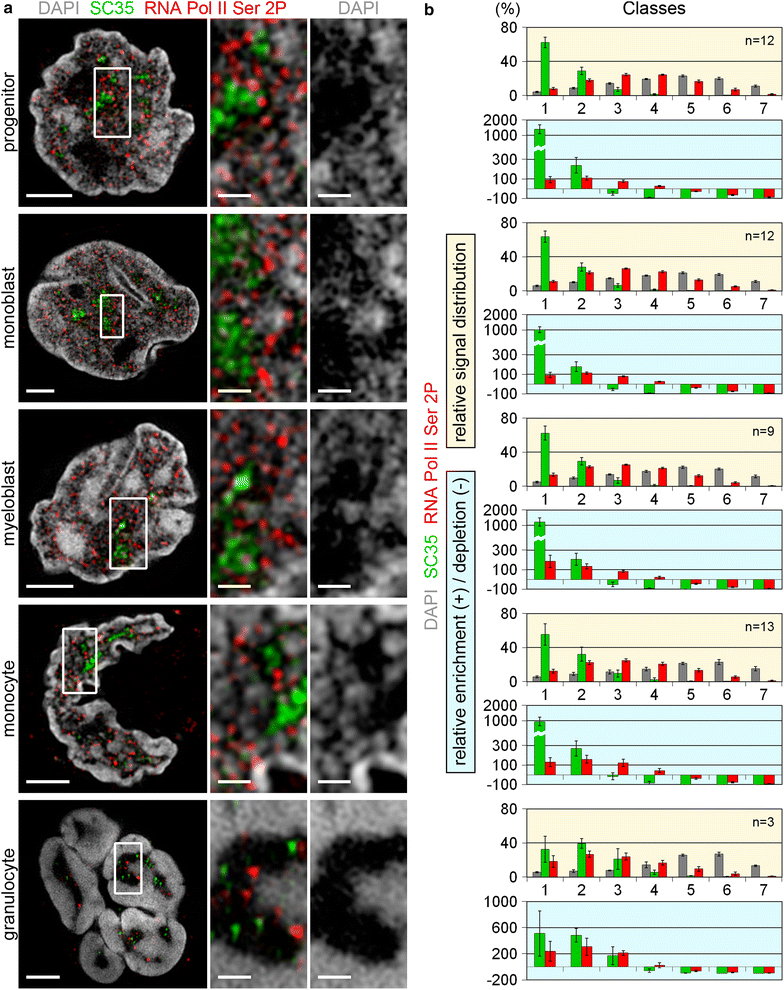
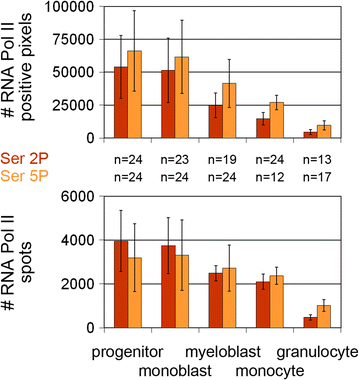
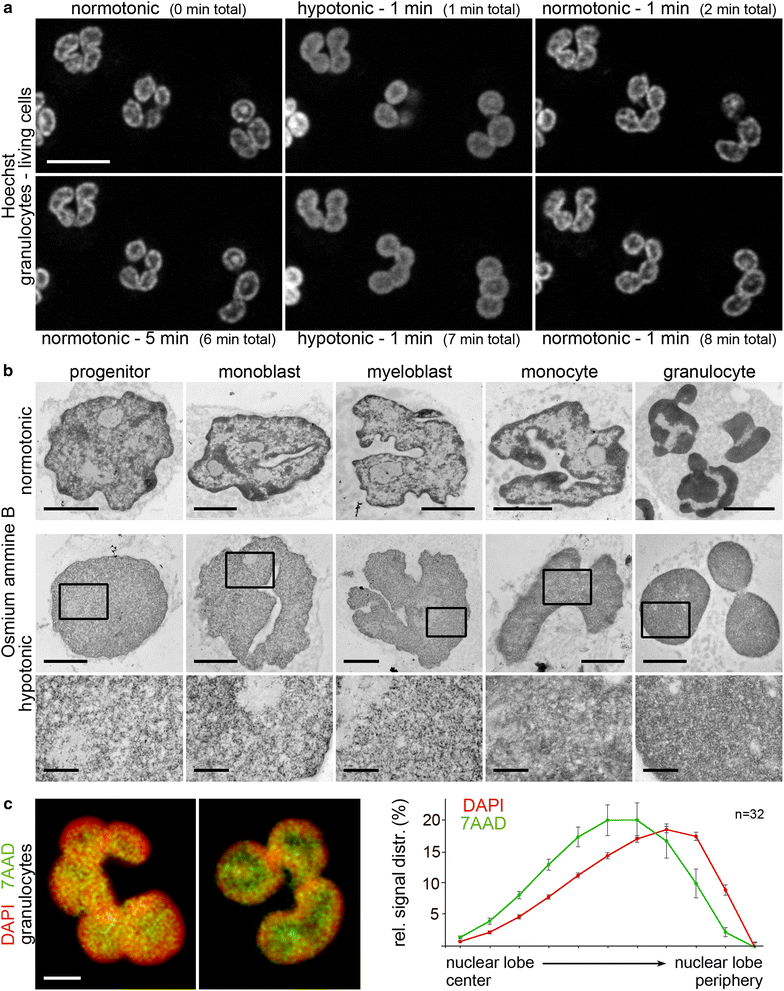
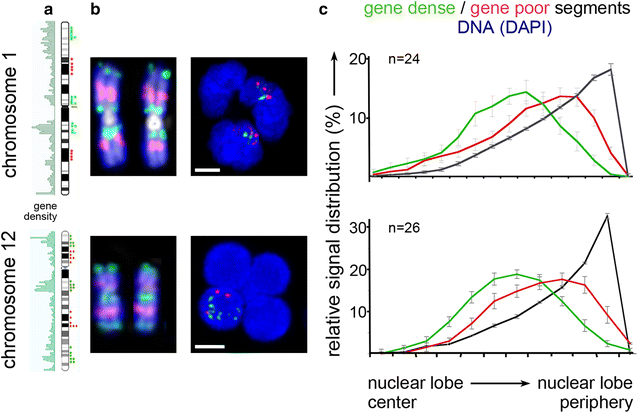
Results: Common human hematopoietic progenitors, myeloid precursor cells, differentiated monocytes and granulocytes analyzed by super-resolution fluorescence microscopy and electron microscopy revealed profound differences with respect to global chromatin arrangements, the nuclear space occupied by the interchromatin compartment and the distribution of nuclear pores. In contrast, we noted a consistent organization in all cell types with regard to two co-aligned networks, an active (ANC) and an inactive (INC) nuclear compartment delineated by functionally relevant hallmarks. The ANC is enriched in active RNA polymerase II, splicing speckles and histone signatures for transcriptionally competent chromatin (H3K4me3), whereas the INC carries marks for repressed chromatin (H3K9me3).
Conclusions: Our findings substantiate the conservation of the recently published ANC-INC network model of mammalian nuclear organization during human myelopoiesis irrespective of profound changes of the global nuclear architecture observed during this differentiation process. According to this model, two spatially co-aligned and functionally interacting active and inactive nuclear compartments (ANC and INC) pervade the nuclear space.
Keywords: Active nuclear compartment; Chromatin density classification; Chromatin domain; Electron microscopy; Interchromatin compartment; Myelopoiesis; Nuclear architecture; Perichromatin region; Somatic cell differentiation; Super-resolution microscopy.
Figures



References
-
- Markaki Y, Gunkel M, Schermelleh L, Beichmanis S, Neumann J, et al. Functional nuclear organization of transcription and DNA replication: a topographical marriage between chromatin domains and the interchromatin compartment. Cold Spring Harb Symp Quant Biol. 2010;75:475–492. doi: 10.1101/sqb.2010.75.042. - DOI - PubMed
-
- Markaki Y, Smeets D, Fiedler S, Schmid VJ, Schermelleh L, et al. The potential of 3D-FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture: 3D structured illumination microscopy of defined chromosomal structures visualized by 3D (immuno)-FISH opens new perspectives for studies of nuclear architecture. Bioessays. 2012;34:412–426. doi: 10.1002/bies.201100176. - DOI - PubMed
-
- Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, et al. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009;17:801–810. doi: 10.1007/s10577-009-9070-x. - DOI - PubMed
-
- Smeets D, Markaki Y, Schmid VJ, Kraus F, Tattermusch A, et al. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin. 2014;7:8. doi: 10.1186/1756-8935-7-8. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

