Putative Breast Cancer Driver Mutations in TBX3 Cause Impaired Transcriptional Repression
- PMID: 26579496
- PMCID: PMC4625211
- DOI: 10.3389/fonc.2015.00244
Putative Breast Cancer Driver Mutations in TBX3 Cause Impaired Transcriptional Repression
Abstract
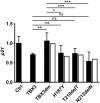
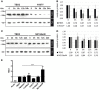
The closely related T-box transcription factors TBX2 and TBX3 are frequently overexpressed in melanoma and various types of human cancers, in particular, breast cancer. The overexpression of TBX2 and TBX3 can have several cellular effects, among them suppression of senescence, promotion of epithelial-mesenchymal transition, and invasive cell motility. In contrast, loss of function of TBX3 and most other human T-box genes causes developmental haploinsufficiency syndromes. Stephens and colleagues (1), by exome sequencing of breast tumor samples, identified five different mutations in TBX3, all affecting the DNA-binding T-domain. One in-frame deletion of a single amino acid, p.N212delN, was observed twice. Due to the clustering of these mutations to the T-domain and for statistical reasons, TBX3 was inferred to be a driver gene in breast cancer. Since mutations in the T-domain generally cause loss of function and because the tumorigenic action of TBX3 has generally been attributed to overexpression, we determined whether the putative driver mutations had loss- or gain-of-function properties. We tested two in-frame deletions, one missense, and one frameshift mutant protein for DNA-binding in vitro, and for target gene repression in cell culture. In addition, we performed an in silico analysis of somatic TBX mutations in breast cancer, collected in The Cancer Genome Atlas (TCGA). Both the experimental and the in silico analysis indicate that the observed mutations predominantly cause loss of TBX3 function.
Keywords: TBX3; breast cancer; driver mutation; frameshift mutation; in-frame deletion; p21; somatic mutations.
Figures

Similar articles
-
A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome.Hum Mol Genet. 2001 Oct 1;10(21):2403-13. doi: 10.1093/hmg/10.21.2403. Hum Mol Genet. 2001. PMID: 11689487
-
The Highly Homologous T-Box Transcription Factors, TBX2 and TBX3, Have Distinct Roles in the Oncogenic Process.Genes Cancer. 2010 Mar;1(3):272-82. doi: 10.1177/1947601910365160. Genes Cancer. 2010. PMID: 21779450 Free PMC article.
-
TBX3 and its splice variant TBX3 + exon 2a are functionally similar.Pigment Cell Melanoma Res. 2008 Jun;21(3):379-87. doi: 10.1111/j.1755-148X.2008.00461.x. Epub 2008 Apr 26. Pigment Cell Melanoma Res. 2008. PMID: 18444963
-
TBX2 and TBX3: the special value for anticancer drug targets.Biochim Biophys Acta. 2010 Dec;1806(2):268-74. doi: 10.1016/j.bbcan.2010.07.001. Epub 2010 Jul 17. Biochim Biophys Acta. 2010. PMID: 20624445 Free PMC article. Review.
-
The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer.J Mammary Gland Biol Neoplasia. 2013 Jun;18(2):143-7. doi: 10.1007/s10911-013-9282-8. Epub 2013 Apr 28. J Mammary Gland Biol Neoplasia. 2013. PMID: 23624936 Free PMC article. Review.
Cited by
-
TBX3 stimulates proliferation and stem cell self-renewal in bladder carcinoma.Histol Histopathol. 2023 Jan;38(1):65-72. doi: 10.14670/HH-18-496. Epub 2022 Jul 20. Histol Histopathol. 2023. PMID: 35856500
-
Mixed ductal-lobular carcinomas: evidence for progression from ductal to lobular morphology.J Pathol. 2018 Apr;244(4):460-468. doi: 10.1002/path.5040. Epub 2018 Mar 9. J Pathol. 2018. PMID: 29344954 Free PMC article.
-
Population-specific Mutation Patterns in Breast Tumors from African American, European American, and Kenyan Patients.Cancer Res Commun. 2023 Nov 7;3(11):2244-2255. doi: 10.1158/2767-9764.CRC-23-0165. Cancer Res Commun. 2023. PMID: 37902422 Free PMC article.
-
A statistical learning method for simultaneous copy number estimation and subclone clustering with single-cell sequencing data.Genome Res. 2024 Feb 7;34(1):85-93. doi: 10.1101/gr.278098.123. Genome Res. 2024. PMID: 38290978 Free PMC article.
-
The T-box transcription factor TBX3 drives proliferation by direct repression of the p21(WAF1) cyclin-dependent kinase inhibitor.Cell Div. 2016 Apr 22;11:6. doi: 10.1186/s13008-016-0019-0. eCollection 2016. Cell Div. 2016. PMID: 27110270 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

