Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation
- PMID: 26579999
- PMCID: PMC11388834
- DOI: 10.1111/pbi.12502
Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation
Abstract
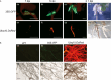
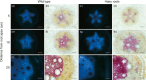
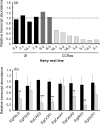
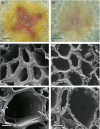
Eucalyptus are of tremendous economic importance being the most planted hardwoods worldwide for pulp and paper, timber and bioenergy. The recent release of the Eucalyptus grandis genome sequence pointed out many new candidate genes potentially involved in secondary growth, wood formation or lineage-specific biosynthetic pathways. Their functional characterization is, however, hindered by the tedious, time-consuming and inefficient transformation systems available hitherto for eucalypts. To overcome this limitation, we developed a fast, reliable and efficient protocol to obtain and easily detect co-transformed E. grandis hairy roots using fluorescent markers, with an average efficiency of 62%. We set up conditions both to cultivate excised roots in vitro and to harden composite plants and verified that hairy root morphology and vascular system anatomy were similar to wild-type ones. We further demonstrated that co-transformed hairy roots are suitable for medium-throughput functional studies enabling, for instance, protein subcellular localization, gene expression patterns through RT-qPCR and promoter expression, as well as the modulation of endogenous gene expression. Down-regulation of the Eucalyptus cinnamoyl-CoA reductase1 (EgCCR1) gene, encoding a key enzyme in lignin biosynthesis, led to transgenic roots with reduced lignin levels and thinner cell walls. This gene was used as a proof of concept to demonstrate that the function of genes involved in secondary cell wall biosynthesis and wood formation can be elucidated in transgenic hairy roots using histochemical, transcriptomic and biochemical approaches. The method described here is timely because it will accelerate gene mining of the genome for both basic research and industry purposes.
Keywords: Agrobacterium rhizogenes; Eucalyptus; hairy roots; lignin; secondary cell wall; xylem.
© 2015 Society for Experimental Biology, Association of Applied Biologists and John Wiley & Sons Ltd.
Figures




References
-
- Alpizar, E. , Dechamp, E. , Espeout, S. , Royer, M. , Lecouls, A.C. , Nicole, M. , Bertrand, B. et al. (2006) Efficient production of Agrobacterium rhizogenes‐transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell Rep. 25, 959–967. - PubMed
-
- Baghdady, A. , Blervacq, A.‐S. , Jouanin, L. , Grima‐Pettenati, J. , Sivadon, P. and Hawkins, S. (2006) Eucalyptus gunnii CCR and CAD2 promoters are active in lignifying cells during primary and secondary xylem formation in Arabidopsis thaliana . Plant Physiol. Biochem. 44, 674–683. - PubMed
-
- Balasubramanian, A. , Venkatachalam, R. , Selvakesavan, K.R. , Mary, A. , Gherbi, H. , Svistoonoff, S. , Franche, C. et al. (2011) Optimisation of methods for Agrobacterium rhizogenes mediated generation of composite plants in Eucalyptus camaldulensis . BMC Proc. 5, O45.
-
- Bécard, G. and Fortin, J.A. (1988) Early events of vesicular‐arbuscular mycorrhiza formation on Ri T‐DNA transformed roots. New Phytol. 108, 211–218. - PubMed
-
- Boisson‐Dernier, A. , Chabaud, M. , Garcia, F. , Bécard, G. , Rosenberg, C. and Barker, D.G. (2001) Agrobacterium rhizogenes‐transformed roots of Medicago truncatula for the study of nitrogen‐fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

