Molecular Characterization of the Elaeis guineensis Medium-Chain Fatty Acid Diacylglycerol Acyltransferase DGAT1-1 by Heterologous Expression in Yarrowia lipolytica
- PMID: 26581109
- PMCID: PMC4651311
- DOI: 10.1371/journal.pone.0143113
Molecular Characterization of the Elaeis guineensis Medium-Chain Fatty Acid Diacylglycerol Acyltransferase DGAT1-1 by Heterologous Expression in Yarrowia lipolytica
Abstract
Diacylglycerol acyltransferases (DGAT) are involved in the acylation of sn-1,2-diacylglycerol. Palm kernel oil, extracted from Elaeis guineensis (oil palm) seeds, has a high content of medium-chain fatty acids mainly lauric acid (C12:0). A putative E. guineensis diacylglycerol acyltransferase gene (EgDGAT1-1) is expressed at the onset of lauric acid accumulation in the seed endosperm suggesting that it is a determinant of medium-chain triacylglycerol storage. To test this hypothesis, we thoroughly characterized EgDGAT1-1 activity through functional complementation of a Yarrowia lipolytica mutant strain devoid of neutral lipids. EgDGAT1-1 expression is sufficient to restore triacylglycerol accumulation in neosynthesized lipid droplets. A comparative functional study with Arabidopsis thaliana DGAT1 highlighted contrasting substrate specificities when the recombinant yeast was cultured in lauric acid supplemented medium. The EgDGAT1-1 expressing strain preferentially accumulated medium-chain triacylglycerols whereas AtDGAT1 expression induced long-chain triacylglycerol storage in Y. lipolytica. EgDGAT1-1 localized to the endoplasmic reticulum where TAG biosynthesis takes place. Reestablishing neutral lipid accumulation in the Y. lipolytica mutant strain did not induce major reorganization of the yeast microsomal proteome. Overall, our findings demonstrate that EgDGAT1-1 is an endoplasmic reticulum DGAT with preference for medium-chain fatty acid substrates, in line with its physiological role in palm kernel. The characterized EgDGAT1-1 could be used to promote medium-chain triacylglycerol accumulation in microbial-produced oil for industrial chemicals and cosmetics.
Conflict of interest statement
Figures





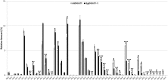
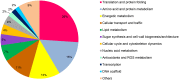

References
-
- Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta. 2000. September 18;1469(2):101–20. - PubMed
-
- Beopoulos A, Haddouche R, Kabran P, Dulermo T, Chardot T, Nicaud JM. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl Microbiol Biotechnol. 2012. February;93(4):1523–37. 10.1007/s00253-011-3506-x - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

