MethylRAD: a simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes
- PMID: 26581575
- PMCID: PMC4680569
- DOI: 10.1098/rsob.150130
MethylRAD: a simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes
Abstract
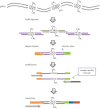
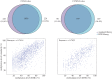
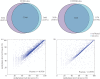
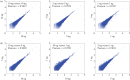
Characterization of dynamic DNA methylomes in diverse phylogenetic groups has attracted growing interest for a better understanding of the evolution of DNA methylation as well as its function and biological significance in eukaryotes. Sequencing-based methods are promising in fulfilling this task. However, none of the currently available methods offers the 'perfect solution', and they have limitations that prevent their application in the less studied phylogenetic groups. The recently discovered Mrr-like enzymes are appealing for new method development, owing to their ability to collect 32-bp methylated DNA fragments from the whole genome for high-throughput sequencing. Here, we have developed a simple and scalable DNA methylation profiling method (called MethylRAD) using Mrr-like enzymes. MethylRAD allows for de novo (reference-free) methylation analysis, extremely low DNA input (e.g. 1 ng) and adjustment of tag density, all of which are still unattainable for most widely used methylation profiling methods such as RRBS and MeDIP. We performed extensive analyses to validate the power and accuracy of our method in both model (plant Arabidopsis thaliana) and non-model (scallop Patinopecten yessoensis) species. We further demonstrated its great utility in identification of a gene (LPCAT1) that is potentially crucial for carotenoid accumulation in scallop adductor muscle. MethylRAD has several advantages over existing tools and fills a void in the current epigenomic toolkit by providing a universal tool that can be used for diverse research applications, e.g. from model to non-model species, from ordinary to precious samples and from small to large genomes, but at an affordable cost.
Keywords: DNA methylation; MRR-like enzyme; MethylRAD; epigenomics.
© 2015 The Authors.
Figures





References
-
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. (doi:10.1038/nrg2719) - DOI - PMC - PubMed
-
- Robertson KD. 2005. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610. (doi:10.1038/nrg1655) - DOI - PubMed
-
- Hirst M, Marra MA. 2011. Next generation sequencing based approaches to epigenomics. Brief Funct. Genomics 9, 455–465. (doi:10.1093/bfgp/elq035) - DOI - PMC - PubMed
-
- Meissner A, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770. (doi:10.1038/nature07107) - DOI - PMC - PubMed
-
- Down TA, et al. 2008. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 26, 779–785. (doi:10.1038/nbt1414) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

