Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype
- PMID: 26582133
- PMCID: PMC4668935
- DOI: 10.1016/j.cell.2015.10.049
Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype
Abstract
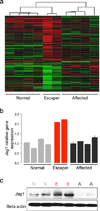
Duchenne muscular dystrophy (DMD), caused by mutations at the dystrophin gene, is the most common form of muscular dystrophy. There is no cure for DMD and current therapeutic approaches to restore dystrophin expression are only partially effective. The absence of dystrophin in muscle results in dysregulation of signaling pathways, which could be targets for disease therapy and drug discovery. Previously, we identified two exceptional Golden Retriever muscular dystrophy (GRMD) dogs that are mildly affected, have functional muscle, and normal lifespan despite the complete absence of dystrophin. Now, our data on linkage, whole-genome sequencing, and transcriptome analyses of these dogs compared to severely affected GRMD and control animals reveals that increased expression of Jagged1 gene, a known regulator of the Notch signaling pathway, is a hallmark of the mild phenotype. Functional analyses demonstrate that Jagged1 overexpression ameliorates the dystrophic phenotype, suggesting that Jagged1 may represent a target for DMD therapy in a dystrophin-independent manner. PAPERCLIP.
Keywords: DMD; Jagged1; dystrophin; genetic modifier; muscle.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. - PubMed
-
- Axelsson E, Ratnakumar A, Arendt ML, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar A, Lindblad-Toh K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. - PubMed
-
- Bassett D, Currie PD. Identification of a zebrafish model of muscular dystrophy. Clinical and experimental pharmacology & physiology. 2004;31:537–540. - PubMed
-
- Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Keenan DG, Currie PD. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–5860. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

