EBI metagenomics in 2016--an expanding and evolving resource for the analysis and archiving of metagenomic data
- PMID: 26582919
- PMCID: PMC4702853
- DOI: 10.1093/nar/gkv1195
EBI metagenomics in 2016--an expanding and evolving resource for the analysis and archiving of metagenomic data
Abstract
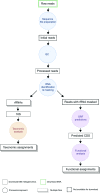
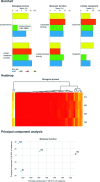
EBI metagenomics (https://www.ebi.ac.uk/metagenomics/) is a freely available hub for the analysis and archiving of metagenomic and metatranscriptomic data. Over the last 2 years, the resource has undergone rapid growth, with an increase of over five-fold in the number of processed samples and consequently represents one of the largest resources of analysed shotgun metagenomes. Here, we report the status of the resource in 2016 and give an overview of new developments. In particular, we describe updates to data content, a complete overhaul of the analysis pipeline, streamlining of data presentation via the website and the development of a new web based tool to compare functional analyses of sequence runs within a study. We also highlight two of the higher profile projects that have been analysed using the resource in the last year: the oceanographic projects Ocean Sampling Day and Tara Oceans.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Pace N.R., Stahl D.A., Lane D.J., Olsen G.J. The analysis of natural microbial populations by ribosomal RNA sequences. In: Marshall K, editor. Advances in Microbial Ecology. Vol. 9. Boston, MA: Springer; 1986. pp. 1–55.
-
- Olsen G.J., Lane D.J., Giovannoni S.J., Pace N.R., Stahl D.A. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 1986;40:337–365. - PubMed
-
- Handelsman J., Rondon M.R., Brady S.F., Clardy J., Goodman R.M. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 1998;5:R245–R249. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

