Protein Destabilization as a Common Factor in Diverse Inherited Disorders
- PMID: 26584803
- PMCID: PMC4712097
- DOI: 10.1007/s00239-015-9717-5
Protein Destabilization as a Common Factor in Diverse Inherited Disorders
Abstract
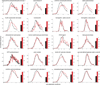
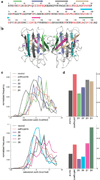
Protein destabilization by amino acid substitutions is proposed to play a prominent role in widespread inherited human disorders, not just those known to involve protein misfolding and aggregation. To test this hypothesis, we computationally evaluate the effects on protein stability of all possible amino acid substitutions in 20 disease-associated proteins with multiple identified pathogenic missense mutations. For 18 of the 20 proteins studied, substitutions at known positions of pathogenic mutations are significantly more likely to destabilize the native protein fold (as indicated by more positive values of ∆∆G). Thus, positions identified as sites of disease-associated mutations, as opposed to non-disease-associated sites, are predicted to be more vulnerable to protein destabilization upon amino acid substitution. This finding supports the notion that destabilization of native protein structure underlies the pathogenicity of broad set of missense mutations, even in cases where reduced protein stability and/or aggregation are not characteristic of the disease state.
Keywords: Aggregation; Destabilization; Inherited disorder; Pathogenic mutation; Stability.
Conflict of interest statement
Figures
References
-
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–1244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

