Proteomics of Primary Cilia by Proximity Labeling
- PMID: 26585297
- PMCID: PMC4662609
- DOI: 10.1016/j.devcel.2015.10.015
Proteomics of Primary Cilia by Proximity Labeling
Abstract
While cilia are recognized as important signaling organelles, the extent of ciliary functions remains unknown because of difficulties in cataloguing proteins from mammalian primary cilia. We present a method that readily captures rapid snapshots of the ciliary proteome by selectively biotinylating ciliary proteins using a cilia-targeted proximity labeling enzyme (cilia-APEX). Besides identifying known ciliary proteins, cilia-APEX uncovered several ciliary signaling molecules. The kinases PKA, AMPK, and LKB1 were validated as bona fide ciliary proteins and PKA was found to regulate Hedgehog signaling in primary cilia. Furthermore, proteomics profiling of Ift27/Bbs19 mutant cilia correctly detected BBSome accumulation inside Ift27(-/-) cilia and revealed that β-arrestin 2 and the viral receptor CAR are candidate cargoes of the BBSome. This work demonstrates that proximity labeling can be applied to proteomics of non-membrane-enclosed organelles and suggests that proteomics profiling of cilia will enable a rapid and powerful characterization of ciliopathies.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
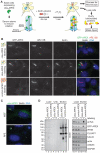

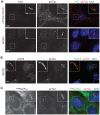
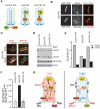
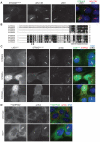
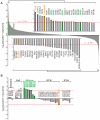
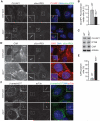
Comment in
-
Taking Snapshots of Ciliary Signaling.Dev Cell. 2015 Nov 23;35(4):399-400. doi: 10.1016/j.devcel.2015.11.006. Dev Cell. 2015. PMID: 26609953
References
-
- Anholt RR. Molecular physiology of olfaction. Am J Physiol. 1989;257:C1043–C1054. - PubMed
-
- Barzi M, Berenguer J, Menendez A, Alvarez-Rodriguez R, Pons S. Sonic-hedgehog-mediated proliferation requires the localization of PKA to the cilium base. J Cell Sci. 2010;123:62–69. - PubMed
-
- Bishop GA, Berbari NF, Lewis J, Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol. 2007;505:562–571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

