Mist1 Expressing Gastric Stem Cells Maintain the Normal and Neoplastic Gastric Epithelium and Are Supported by a Perivascular Stem Cell Niche
- PMID: 26585400
- PMCID: PMC4684751
- DOI: 10.1016/j.ccell.2015.10.003
Mist1 Expressing Gastric Stem Cells Maintain the Normal and Neoplastic Gastric Epithelium and Are Supported by a Perivascular Stem Cell Niche
Abstract
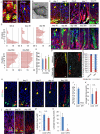
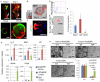
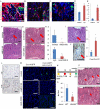
The regulation and stem cell origin of normal and neoplastic gastric glands are uncertain. Here, we show that Mist1 expression marks quiescent stem cells in the gastric corpus isthmus. Mist1(+) stem cells serve as a cell-of-origin for intestinal-type cancer with the combination of Kras and Apc mutation and for diffuse-type cancer with the loss of E-cadherin. Diffuse-type cancer development is dependent on inflammation mediated by Cxcl12(+) endothelial cells and Cxcr4(+) gastric innate lymphoid cells (ILCs). These cells form the perivascular gastric stem cell niche, and Wnt5a produced from ILCs activates RhoA to inhibit anoikis in the E-cadherin-depleted cells. Targeting Cxcr4, ILCs, or Wnt5a inhibits diffuse-type gastric carcinogenesis, providing targets within the neoplastic gastric stem cell niche.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Lifting the Mist on Gastric Stem Cells.Cell Stem Cell. 2016 Jan 7;18(1):7-9. doi: 10.1016/j.stem.2015.12.008. Cell Stem Cell. 2016. PMID: 26748749
References
-
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. - PubMed
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

