Expansion of Interstitial Telomeric Sequences in Yeast
- PMID: 26586439
- PMCID: PMC4662878
- DOI: 10.1016/j.celrep.2015.10.023
Expansion of Interstitial Telomeric Sequences in Yeast
Abstract
Telomeric repeats located within chromosomes are called interstitial telomeric sequences (ITSs). They are polymorphic in length and are likely hotspots for initiation of chromosomal rearrangements that have been linked to human disease. Using our S. cerevisiae system to study repeat-mediated genome instability, we have previously shown that yeast telomeric (Ytel) repeats induce various gross chromosomal rearrangements (GCR) when their G-rich strands serve as the lagging strand template for replication (G orientation). Here, we show that interstitial Ytel repeats in the opposite C orientation prefer to expand rather than cause GCR. A tract of eight Ytel repeats expands at a rate of 4 × 10(-4) per replication, ranking them among the most expansion-prone DNA microsatellites. A candidate-based genetic analysis implicates both post-replication repair and homologous recombination pathways in the expansion process. We propose a model for Ytel repeat expansions and discuss its applications for genome instability and alternative telomere lengthening (ALT).
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
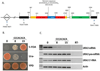

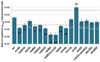
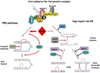
References
-
- Azzalin CM, Mucciolo E, Bertoni L, Giulotto E. Fluorescence in situ hybridization with a synthetic (T2AG3)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet Cell Genet. 1997;78:112–115. - PubMed
-
- Azzalin CM, Nergadze SG, Giulotto E. Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma. 2001;110:75–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

