Metabolic and genomic analysis elucidates strain-level variation in Microbacterium spp. isolated from chromate contaminated sediment
- PMID: 26587353
- PMCID: PMC4647564
- DOI: 10.7717/peerj.1395
Metabolic and genomic analysis elucidates strain-level variation in Microbacterium spp. isolated from chromate contaminated sediment
Abstract
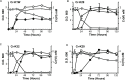
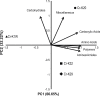
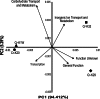
Hexavalent chromium [Cr(VI)] is a soluble carcinogen that has caused widespread contamination of soil and water in many industrial nations. Bacteria have the potential to aid remediation as certain strains can catalyze the reduction of Cr(VI) to insoluble and less toxic Cr(III). Here, we examine Cr(VI) reducing Microbacterium spp. (Cr-K1W, Cr-K20, Cr-K29, and Cr-K32) isolated from contaminated sediment (Seymore, Indiana) and show varying chromate responses despite the isolates' phylogenetic similarity (i.e., identical 16S rRNA gene sequences). Detailed analysis identified differences based on genomic metabolic potential, growth and general metabolic capabilities, and capacity to resist and reduce Cr(VI). Taken together, the discrepancies between the isolates demonstrate the complexity inter-strain variation can have on microbial physiology and related biogeochemical processes.
Keywords: Chromium reduction; Microbacterium; Strain-level variation.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
References
-
- Ackerley DF, Gonzalez CF, Keyhan M, Blake R, Matin A. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environmental Microbiology. 2004a;6:851–860. doi: 10.1111/j.1462-2920.2004.00639.x. - DOI - PubMed
-
- Bååth E. Effects of heavy metals in soil on microbial processes and populations (a review) Water, Air, and Soil Pollution. 1989;47:335–379. doi: 10.1007/BF00279331. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

