Global Analysis of the Fungal Microbiome in Cystic Fibrosis Patients Reveals Loss of Function of the Transcriptional Repressor Nrg1 as a Mechanism of Pathogen Adaptation
- PMID: 26588216
- PMCID: PMC4654494
- DOI: 10.1371/journal.ppat.1005308
Global Analysis of the Fungal Microbiome in Cystic Fibrosis Patients Reveals Loss of Function of the Transcriptional Repressor Nrg1 as a Mechanism of Pathogen Adaptation
Abstract
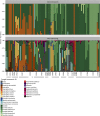
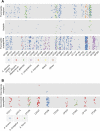
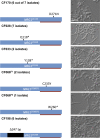
The microbiome shapes diverse facets of human biology and disease, with the importance of fungi only beginning to be appreciated. Microbial communities infiltrate diverse anatomical sites as with the respiratory tract of healthy humans and those with diseases such as cystic fibrosis, where chronic colonization and infection lead to clinical decline. Although fungi are frequently recovered from cystic fibrosis patient sputum samples and have been associated with deterioration of lung function, understanding of species and population dynamics remains in its infancy. Here, we coupled high-throughput sequencing of the ribosomal RNA internal transcribed spacer 1 (ITS1) with phenotypic and genotypic analyses of fungi from 89 sputum samples from 28 cystic fibrosis patients. Fungal communities defined by sequencing were concordant with those defined by culture-based analyses of 1,603 isolates from the same samples. Different patients harbored distinct fungal communities. There were detectable trends, however, including colonization with Candida and Aspergillus species, which was not perturbed by clinical exacerbation or treatment. We identified considerable inter- and intra-species phenotypic variation in traits important for host adaptation, including antifungal drug resistance and morphogenesis. While variation in drug resistance was largely between species, striking variation in morphogenesis emerged within Candida species. Filamentation was uncoupled from inducing cues in 28 Candida isolates recovered from six patients. The filamentous isolates were resistant to the filamentation-repressive effects of Pseudomonas aeruginosa, implicating inter-kingdom interactions as the selective force. Genome sequencing revealed that all but one of the filamentous isolates harbored mutations in the transcriptional repressor NRG1; such mutations were necessary and sufficient for the filamentous phenotype. Six independent nrg1 mutations arose in Candida isolates from different patients, providing a poignant example of parallel evolution. Together, this combined clinical-genomic approach provides a high-resolution portrait of the fungal microbiome of cystic fibrosis patient lungs and identifies a genetic basis of pathogen adaptation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

