Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development
- PMID: 26588779
- PMCID: PMC4654992
- DOI: 10.1016/j.immuni.2015.11.001
Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development
Abstract
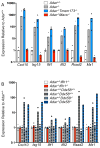
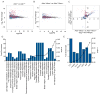
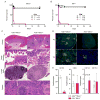
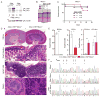
Mutations in ADAR, which encodes the ADAR1 RNA-editing enzyme, cause Aicardi-Goutières syndrome (AGS), a severe autoimmune disease associated with an aberrant type I interferon response. How ADAR1 prevents autoimmunity remains incompletely defined. Here, we demonstrate that ADAR1 is a specific and essential negative regulator of the MDA5-MAVS RNA sensing pathway. Moreover, we uncovered a MDA5-MAVS-independent function for ADAR1 in the development of multiple organs. We showed that the p150 isoform of ADAR1 uniquely regulated the MDA5 pathway, whereas both the p150 and p110 isoforms contributed to development. Abrupt deletion of ADAR1 in adult mice revealed that both of these functions were required throughout life. Our findings delineate genetically separable roles for both ADAR1 isoforms in vivo, with implications for the human diseases caused by ADAR mutations.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol. 2014;192:5993–5997. - PubMed
-
- Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol. 1984;15:49–54. - PubMed
-
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. American journal of medical genetics Part A. 2015;167A:296–312. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

