Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta
- PMID: 26588991
- PMCID: PMC4691893
- DOI: 10.1101/gad.269050.115
Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta
Erratum in
-
Corrigendum: Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta.Genes Dev. 2016 Jan 1;30(1):132. Genes Dev. 2016. PMID: 26728558 Free PMC article. No abstract available.
Abstract
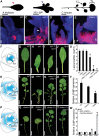
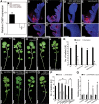
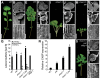
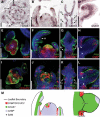
Two interrelated problems in biology are understanding the regulatory logic and predictability of morphological evolution. Here, we studied these problems by comparing Arabidopsis thaliana, which has simple leaves, and its relative, Cardamine hirsuta, which has dissected leaves comprising leaflets. By transferring genes between the two species, we provide evidence for an inverse relationship between the pleiotropy of SHOOTMERISTEMLESS (STM) and BREVIPEDICELLUS (BP) homeobox genes and their ability to modify leaf form. We further show that cis-regulatory divergence of BP results in two alternative configurations of the genetic networks controlling leaf development. In C. hirsuta, ChBP is repressed by the microRNA164A (MIR164A)/ChCUP-SHAPED COTYLEDON (ChCUC) module and ChASYMMETRIC LEAVES1 (ChAS1), thus creating cross-talk between MIR164A/CUC and AS1 that does not occur in A. thaliana. These different genetic architectures lead to divergent interactions of network components and growth regulation in each species. We suggest that certain regulatory genes with low pleiotropy are predisposed to readily integrate into or disengage from conserved genetic networks influencing organ geometry, thus rapidly altering their properties and contributing to morphological divergence.
Keywords: CUP-SHAPED COTYLEDON; Cardamine hirsuta; KNOXI genes; compound leaf; pleiotropy; regulatory evolution.
© 2015 Rast-Somssich et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Aguilar-Martinez JA, Uchida N, Townsley B, West DA, Yanez A, Lynn N, Kimura S, Sinha N. 2015. Transcriptional, posttranscriptional, and posttranslational regulation of SHOOT MERISTEMLESS gene expression in Arabidopsis determines gene function in the shoot apex. Plant Physiol 167: 424–442. - PMC - PubMed
-
- Arnoult L, Su KF, Manoel D, Minervino C, Magrina J, Gompel N, Prud'homme B. 2013. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339: 1423–1426. - PubMed
-
- Arthur W. 2002. The emerging conceptual framework of evolutionary developmental biology. Nature 415: 757–764. - PubMed
-
- Arthur W. 2004. Biased embryos and evolution. University Press, Cambridge, UK.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
