Evidence for Epigenetic Regulation of Pro-Inflammatory Cytokines, Interleukin-12 and Interferon Gamma, in Peripheral Blood Mononuclear Cells from PTSD Patients
- PMID: 26589234
- PMCID: PMC4746110
- DOI: 10.1007/s11481-015-9643-8
Evidence for Epigenetic Regulation of Pro-Inflammatory Cytokines, Interleukin-12 and Interferon Gamma, in Peripheral Blood Mononuclear Cells from PTSD Patients
Abstract
While Post Traumatic Stress Disorder (PTSD) is associated with immune dysfunction, the underlying mechanisms remain unclear. Studies suggest a role for involvement of epigenetic mechanisms and microRNAs (miRNAs). Here, we examined genome-wide histone and DNA methylation in the peripheral blood mononuclear cells (PBMCs) in PTSD. We noted significant differences in histone H3 trimethylation at K4, K9, K27 and K36 sites in PTSD when compared to control. While overall DNA methylation level did not differ significantly between control and PTSD, the promoters of several individual genes (e.g., Interferon gamma (IFNG) and Interleukin (IL)-12B) were differentially methylated. ChIP-seq data revealed that the promoter of IFNG and TBX-21 was associated with the activation marker H3K4me3 in PTSD. The transcript levels of both IFNG and TBX-21 were higher in PTSD correlating well with the altered methylation patterns. Furthermore, PTSD patients showed increased expression of IL-12 in their PBMCs. Analysis of both histone and DNA methylation markers suggested that the expression of IL-12 was also possibly activated through epigenetic modification. Knockdown of lysine (K)-specific demethylase 5B (KDM5B), or inhibition of DNA (Cytosine-5-)-methyltransferase 1 (DNMT1) caused up-regulation of IL-12. Furthermore, the expression of these cytokines was also regulated by miRNAs. Our miRNA microarray identified many downregulated miRNAs in PTSD that are predicted to target IFNG and IL-12. Consequently, we showed that up-regulation of hsa-miR-193a-5p could decrease the expression of IL-12. Overall, the current study demonstrated that the elevated expression of pro-inflammatory cytokines in PTSD patients might be regulated by multiple epigenetic mechanisms and miRNAs.
Keywords: DNA methylation; Histone modification; IL-12; Inflammation; Post-traumatic stress disorder; miRNA.
Figures
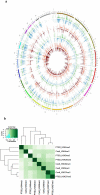


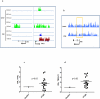



References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004 Sep 16;431(7006):350–5. doi:10.1038/nature02871. - PubMed
-
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi:10.1074/jbc.M500796200. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi:10.1016/S0092-8674(04)00045-5. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi:10.1016/j.cell.2006.02.041. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AT006888/AT/NCCIH NIH HHS/United States
- R01ES019313/ES/NIEHS NIH HHS/United States
- P20GM103641/GM/NIGMS NIH HHS/United States
- R01 MH094755/MH/NIMH NIH HHS/United States
- R01MH094755/MH/NIMH NIH HHS/United States
- R01 ES019313/ES/NIEHS NIH HHS/United States
- P20 GM103641/GM/NIGMS NIH HHS/United States
- P01 AT003961/AT/NCCIH NIH HHS/United States
- R01 AT006888/AT/NCCIH NIH HHS/United States
- P01AT003961/AT/NCCIH NIH HHS/United States
- R01 AI129788/AI/NIAID NIH HHS/United States
- I01 BX001357/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

