Large-scale machine learning for metagenomics sequence classification
- PMID: 26589281
- PMCID: PMC4896366
- DOI: 10.1093/bioinformatics/btv683
Large-scale machine learning for metagenomics sequence classification
Abstract
Motivation: Metagenomics characterizes the taxonomic diversity of microbial communities by sequencing DNA directly from an environmental sample. One of the main challenges in metagenomics data analysis is the binning step, where each sequenced read is assigned to a taxonomic clade. Because of the large volume of metagenomics datasets, binning methods need fast and accurate algorithms that can operate with reasonable computing requirements. While standard alignment-based methods provide state-of-the-art performance, compositional approaches that assign a taxonomic class to a DNA read based on the k-mers it contains have the potential to provide faster solutions.
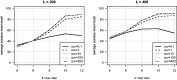
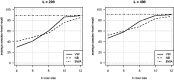
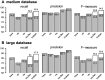
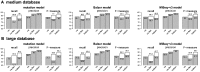
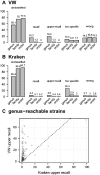
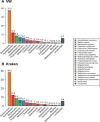
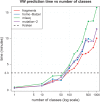
Results: We propose a new rank-flexible machine learning-based compositional approach for taxonomic assignment of metagenomics reads and show that it benefits from increasing the number of fragments sampled from reference genome to tune its parameters, up to a coverage of about 10, and from increasing the k-mer size to about 12. Tuning the method involves training machine learning models on about 10(8) samples in 10(7) dimensions, which is out of reach of standard softwares but can be done efficiently with modern implementations for large-scale machine learning. The resulting method is competitive in terms of accuracy with well-established alignment and composition-based tools for problems involving a small to moderate number of candidate species and for reasonable amounts of sequencing errors. We show, however, that machine learning-based compositional approaches are still limited in their ability to deal with problems involving a greater number of species and more sensitive to sequencing errors. We finally show that the new method outperforms the state-of-the-art in its ability to classify reads from species of lineage absent from the reference database and confirm that compositional approaches achieve faster prediction times, with a gain of 2-17 times with respect to the BWA-MEM short read mapper, depending on the number of candidate species and the level of sequencing noise.
Availability and implementation: Data and codes are available at http://cbio.ensmp.fr/largescalemetagenomics
Contact: pierre.mahe@biomerieux.com
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
References
-
- Agarwal A., et al. (2014) A reliable effective terascale linear learning system. J. Mach. Learn. Res., 15, 1111–1133.
-
- Beygelzimer A., et al. (2009) Error-correcting tournaments. Algorithmic Learn. Theory, 5809, 247–262.
-
- Bottou L. (1998) Online learning and stochastic approximations. Online Learn. Neural Netw., 17, 9–42.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

