Pathobiochemical signatures of cholestatic liver disease in bile duct ligated mice
- PMID: 26589287
- PMCID: PMC4654904
- DOI: 10.1186/s12918-015-0229-0
Pathobiochemical signatures of cholestatic liver disease in bile duct ligated mice
Abstract
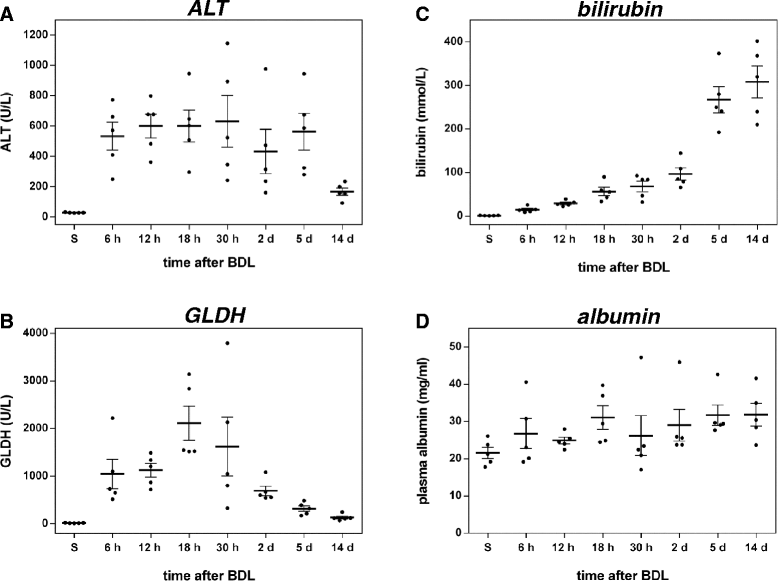
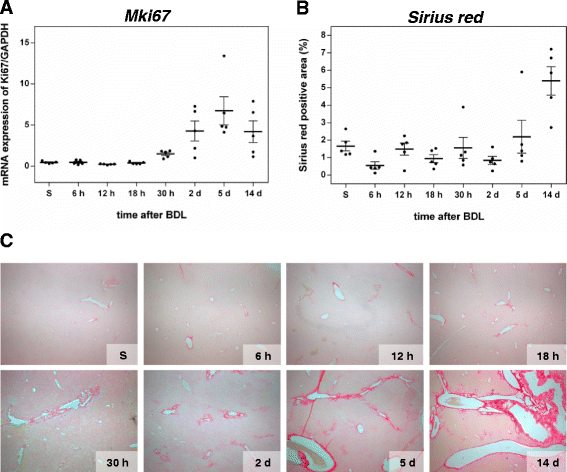
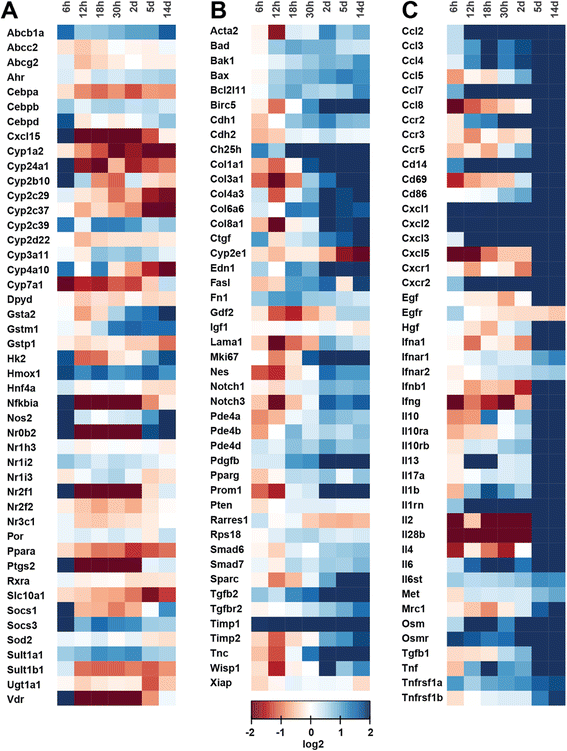
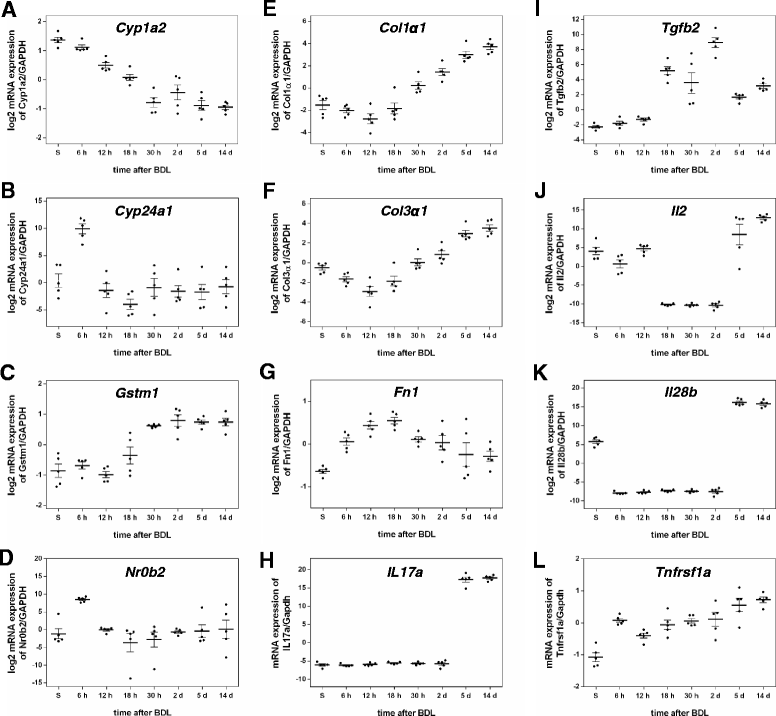
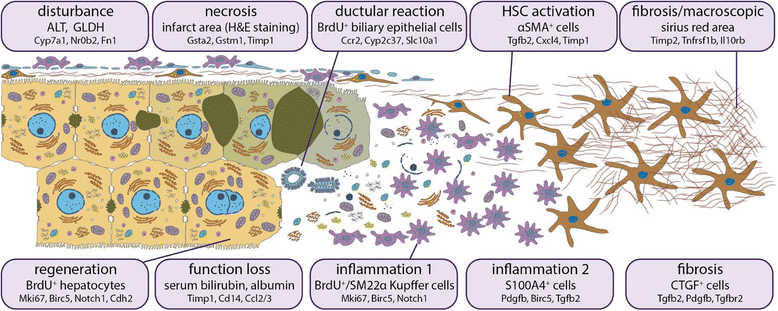
Background: Disrupted bile secretion leads to liver damage characterized by inflammation, fibrosis, eventually cirrhosis, and hepatocellular cancer. As obstructive cholestasis often progresses insidiously, markers for the diagnosis and staging of the disease are urgently needed. To this end, we compiled a comprehensive data set of serum markers, histological parameters and transcript profiles at 8 time points of disease progression after bile duct ligation (BDL) in mice, aiming at identifying a set of parameters that could be used as robust biomarkers for transition of different disease progression phases.
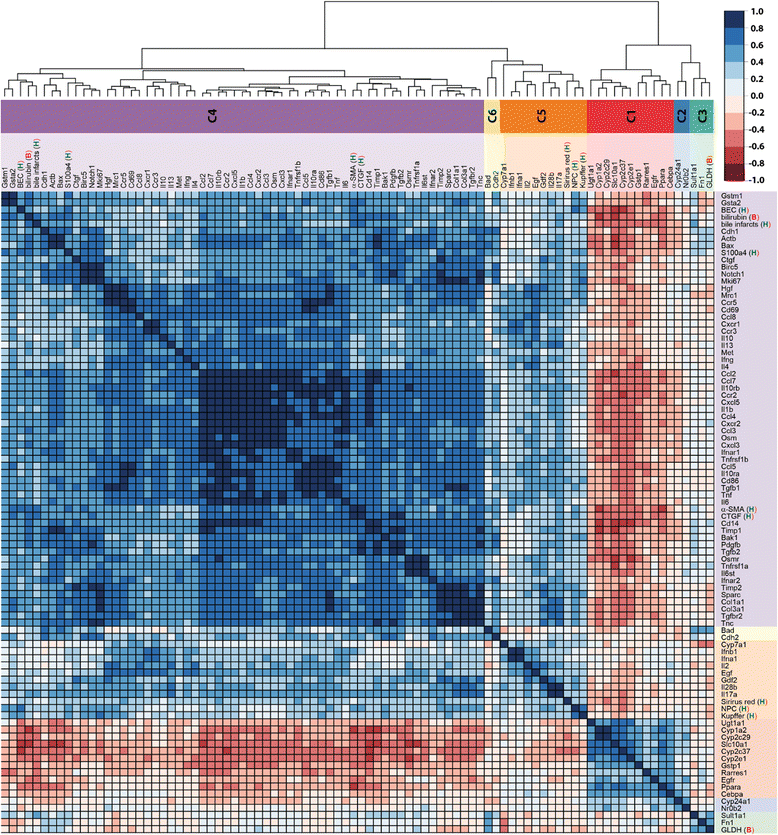
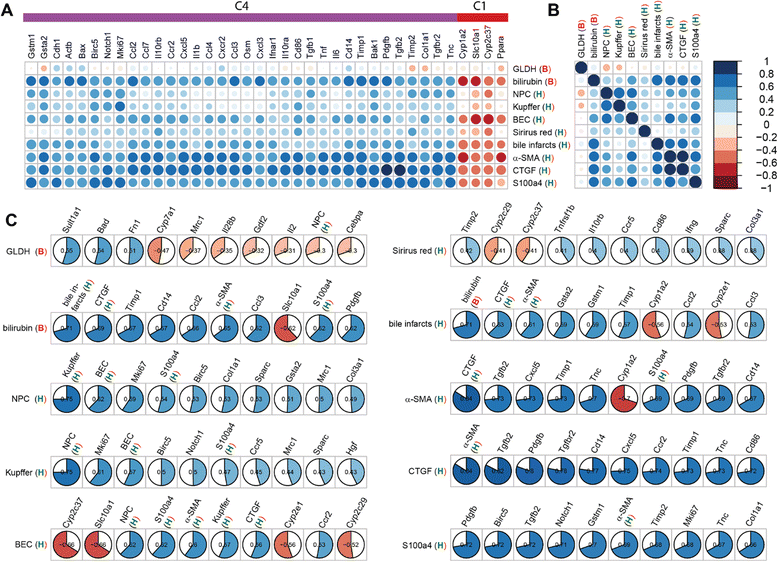
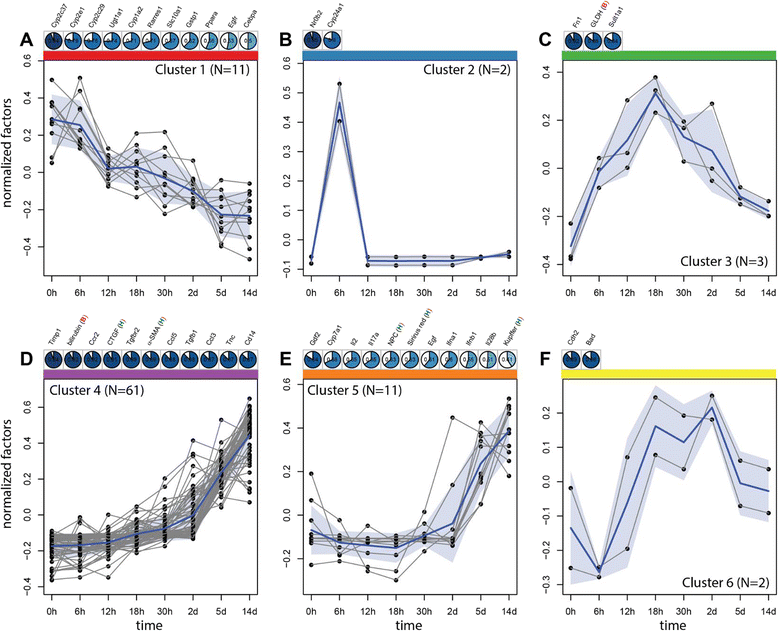
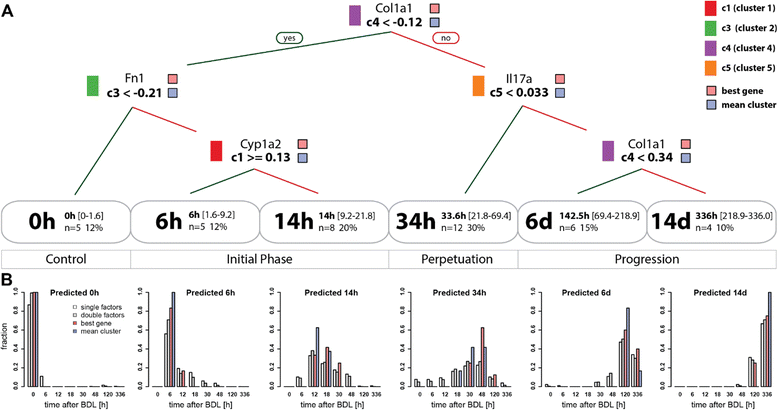
Results: Statistical analysis of the more than 6,000 data points revealed distinct temporal phases of disease. Time course correlation analysis of biochemical, histochemical and mRNA transcript parameters (=factors) defined 6 clusters for different phases of disease progression. The number of CTGF-positive cells provided the most reliable overall measure for disease progression at histological level, bilirubin at biochemical level, and metalloproteinase inhibitor 1 (Timp1) at transcript level. Prominent molecular events exhibited by strong transcript peaks are found for the transcriptional regulator Nr0b2 (Shp) and 1,25-dihydroxyvitamin D(3) 24-hydroxylase (Cyp24a1) at 6 h. Based on these clusters, we constructed a decision tree of factor combinations potentially useful as markers for different time intervals of disease progression. Best prediction for onset of disease is achieved by fibronectin (Fn1), for early disease phase by Cytochrome P450 1A2 (Cyp1a2), passage to perpetuation phase by collagen1α-1 (Col1a1), and transition to the progression phase by interleukin 17-a (Il17a), with early and late progression separated by Col1a1. Notably, these predictions remained stable even for randomly chosen small sub-sets of factors selected from the clusters.
Conclusion: Our detailed time-resolved explorative study of liver homogenates following BDL revealed a well-coordinated response, resulting in disease phase dependent parameter modulations at morphological, biochemical, metabolic and gene expression levels. Interestingly, a small set of selected parameters can be used as diagnostic markers to predict disease stages in mice with cholestatic liver disease.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

