Patterns of gene expression and DNA methylation in human fetal and adult liver
- PMID: 26589361
- PMCID: PMC4654795
- DOI: 10.1186/s12864-015-2066-3
Patterns of gene expression and DNA methylation in human fetal and adult liver
Abstract
Background: DNA methylation is an important epigenetic control mechanism that has been shown to be associated with gene silencing through the course of development, maturation and aging. However, only limited data are available regarding the relationship between methylation and gene expression in human development.
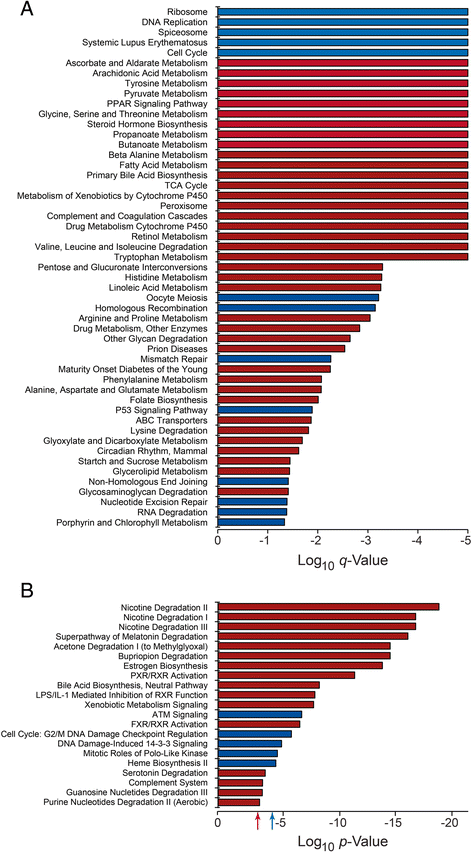
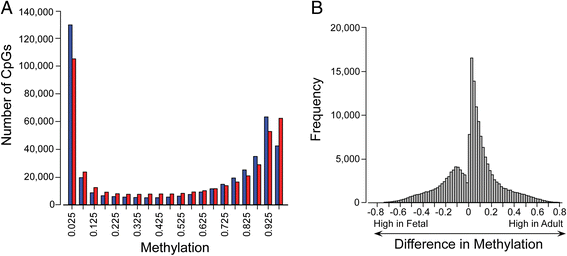
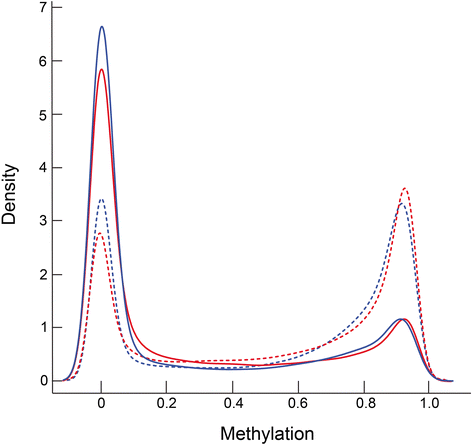
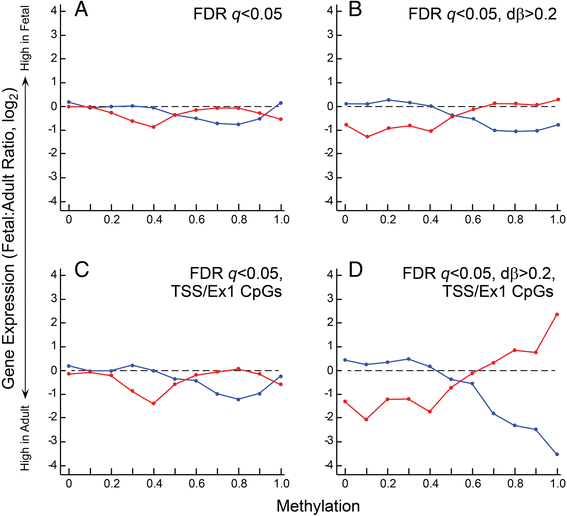
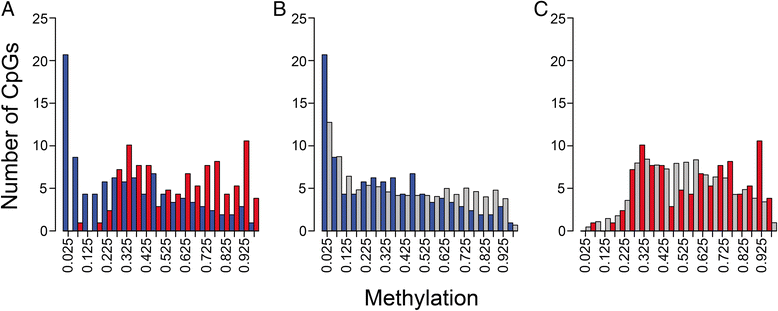
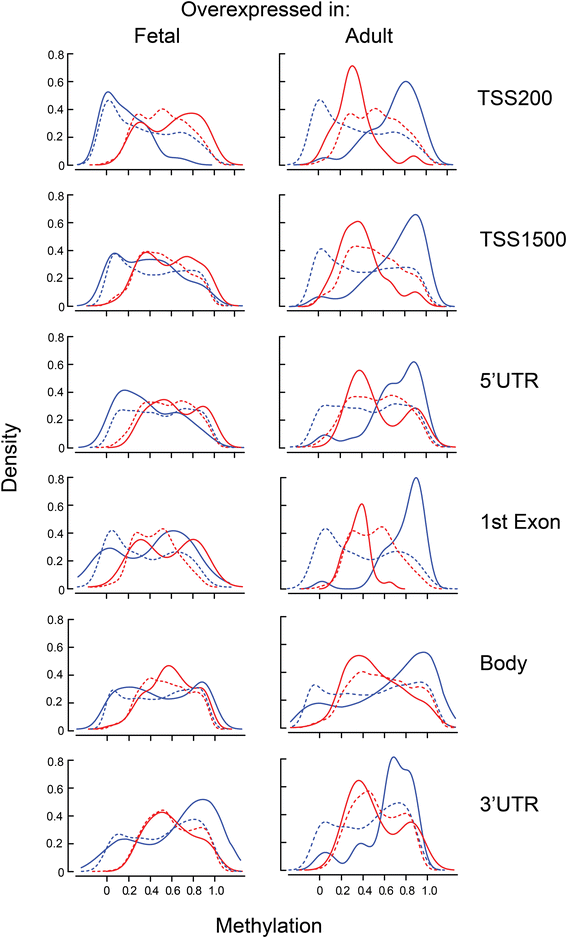
Results: We analyzed the methylome and transcriptome of three human fetal liver samples (gestational age 20-22 weeks) and three adult human liver samples. Genes whose expression differed between fetal and adult numbered 7,673. Adult overexpression was associated with metabolic pathways and, in particular, cytochrome P450 enzymes while fetal overexpression reflected enrichment for DNA replication and repair. Analysis for DNA methylation using the Illumina Infinium 450 K HumanMethylation BeadChip showed that 42% of the quality filtered 426,154 methylation sites differed significantly between adult and fetal tissue (q ≤ 0.05). Differences were small; 69% of the significant sites differed in their mean methylation beta value by ≤0.2. There was a trend among all sites toward higher methylation in the adult samples with the most frequent difference in beta being 0.1. Characterization of the relationship between methylation and expression revealed a clear difference between fetus and adult. Methylation of genes overexpressed in fetal liver showed the same pattern as seen for genes that were similarly expressed in fetal and adult liver. In contrast, adult overexpressed genes showed fetal hypermethylation that differed from the similarly expressed genes. An examination of gene region-specific methylation showed that sites proximal to the transcription start site or within the first exon with a significant fetal-adult difference in beta (>0.2) showed an inverse relationship with gene expression.
Conclusions: Nearly half of the CpGs in human liver show a significant difference in methylation comparing fetal and adult samples. Sites proximal to the transcription start site or within the first exon that show a transition from hypermethylation in the fetus to hypomethylation or intermediate methylation in the adult are associated with inverse changes in gene expression. In contrast, increases in methylation going from fetal to adult are not associated with fetal-to-adult decreased expression. These findings indicate fundamentally different roles for and/or regulation of DNA methylation in human fetal and adult liver.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

