Practical guidelines for B-cell receptor repertoire sequencing analysis
- PMID: 26589402
- PMCID: PMC4654805
- DOI: 10.1186/s13073-015-0243-2
Practical guidelines for B-cell receptor repertoire sequencing analysis
Abstract
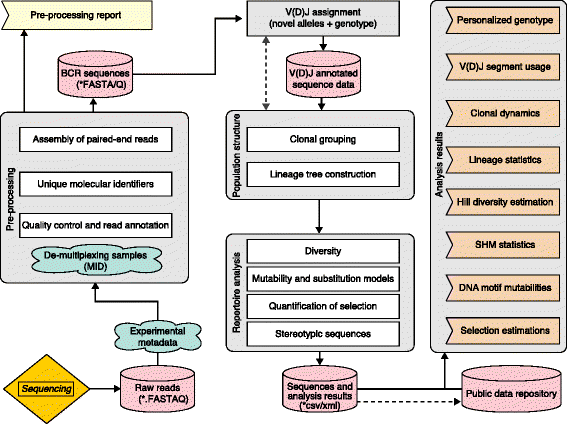
High-throughput sequencing of B-cell immunoglobulin repertoires is increasingly being applied to gain insights into the adaptive immune response in healthy individuals and in those with a wide range of diseases. Recent applications include the study of autoimmunity, infection, allergy, cancer and aging. As sequencing technologies continue to improve, these repertoire sequencing experiments are producing ever larger datasets, with tens- to hundreds-of-millions of sequences. These data require specialized bioinformatics pipelines to be analyzed effectively. Numerous methods and tools have been developed to handle different steps of the analysis, and integrated software suites have recently been made available. However, the field has yet to converge on a standard pipeline for data processing and analysis. Common file formats for data sharing are also lacking. Here we provide a set of practical guidelines for B-cell receptor repertoire sequencing analysis, starting from raw sequencing reads and proceeding through pre-processing, determination of population structure, and analysis of repertoire properties. These include methods for unique molecular identifiers and sequencing error correction, V(D)J assignment and detection of novel alleles, clonal assignment, lineage tree construction, somatic hypermutation modeling, selection analysis, and analysis of stereotyped or convergent responses. The guidelines presented here highlight the major steps involved in the analysis of B-cell repertoire sequencing data, along with recommendations on how to avoid common pitfalls.
Figures


References
-
- Boyd SD, Joshi SA. High-throughput DNA sequencing analysis of antibody repertoires. Microbiol Spectr. 2014;2. doi: 10.1128/microbiolspec.AID-0017-20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

