Molecular mechanism of positive allosteric modulation of the metabotropic glutamate receptor 2 by JNJ-46281222
- PMID: 26589404
- PMCID: PMC4728424
- DOI: 10.1111/bph.13390
Molecular mechanism of positive allosteric modulation of the metabotropic glutamate receptor 2 by JNJ-46281222
Abstract
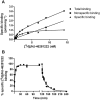
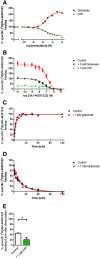
Background and purpose: Allosteric modulation of the mGlu2 receptor is a potential strategy for treatment of various neurological and psychiatric disorders. Here, we describe the in vitro characterization of the mGlu2 positive allosteric modulator (PAM) JNJ-46281222 and its radiolabelled counterpart [(3) H]-JNJ-46281222. Using this novel tool, we also describe the allosteric effect of orthosteric glutamate binding and the presence of a bound G protein on PAM binding and use computational approaches to further investigate the binding mode.
Experimental approach: We have used radioligand binding studies, functional assays, site-directed mutagenesis, homology modelling and molecular dynamics to study the binding of JNJ-46281222.
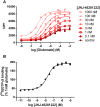
Key results: JNJ-46281222 is an mGlu2 -selective, highly potent PAM with nanomolar affinity (KD = 1.7 nM). Binding of [(3) H]-JNJ-46281222 was increased by the presence of glutamate and greatly reduced by the presence of GTP, indicating the preference for a G protein bound state of the receptor for PAM binding. Its allosteric binding site was visualized and analysed by a computational docking and molecular dynamics study. The simulations revealed amino acid movements in regions expected to be important for activation. The binding mode was supported by [(3) H]-JNJ-46281222 binding experiments on mutant receptors.
Conclusion and implications: Our results obtained with JNJ-46281222 in unlabelled and tritiated form further contribute to our understanding of mGlu2 allosteric modulation. The computational simulations and mutagenesis provide a plausible binding mode with indications of how the ligand permits allosteric activation. This study is therefore of interest for mGlu2 and class C receptor drug discovery.
© 2015 The British Pharmacological Society.
Figures


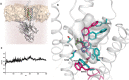

Similar articles
-
Pharmacological characterization of JNJ-40068782, a new potent, selective, and systemically active positive allosteric modulator of the mGlu2 receptor and its radioligand [3H]JNJ-40068782.J Pharmacol Exp Ther. 2013 Sep;346(3):514-27. doi: 10.1124/jpet.113.204990. Epub 2013 Jun 13. J Pharmacol Exp Ther. 2013. PMID: 23766542
-
Pharmacological and molecular characterization of the positive allosteric modulators of metabotropic glutamate receptor 2.Neuropharmacology. 2016 Dec;111:253-265. doi: 10.1016/j.neuropharm.2016.08.032. Epub 2016 Aug 30. Neuropharmacology. 2016. PMID: 27590915 Review.
-
mGlu2 Receptor Agonism, but Not Positive Allosteric Modulation, Elicits Rapid Tolerance towards Their Primary Efficacy on Sleep Measures in Rats.PLoS One. 2015 Dec 11;10(12):e0144017. doi: 10.1371/journal.pone.0144017. eCollection 2015. PLoS One. 2015. PMID: 26658273 Free PMC article.
-
Molecular Switches of Allosteric Modulation of the Metabotropic Glutamate 2 Receptor.Structure. 2017 Jul 5;25(7):1153-1162.e4. doi: 10.1016/j.str.2017.05.021. Epub 2017 Jun 22. Structure. 2017. PMID: 28648611
-
Reprint of Pharmacological and molecular characterization of the positive allosteric modulators of metabotropic glutamate receptor 2.Neuropharmacology. 2017 Mar 15;115:115-127. doi: 10.1016/j.neuropharm.2016.08.040. Epub 2017 Feb 16. Neuropharmacology. 2017. PMID: 28216000 Review.
Cited by
-
Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor.Elife. 2019 Jun 7;8:e45116. doi: 10.7554/eLife.45116. Elife. 2019. PMID: 31172948 Free PMC article.
-
Differential Pharmacology and Binding of mGlu2 Receptor Allosteric Modulators.Mol Pharmacol. 2018 May;93(5):526-540. doi: 10.1124/mol.117.110114. Epub 2018 Mar 15. Mol Pharmacol. 2018. PMID: 29545267 Free PMC article.
-
Identification of the Candidate mGlu2 Allosteric Modulator THRX-195518 through In Silico Method and Evaluation of Its Neuroprotective Potential against Glutamate-Induced Neurotoxicity in SH-SY5Y Cell Line.Curr Issues Mol Biol. 2024 Jan 17;46(1):788-807. doi: 10.3390/cimb46010051. Curr Issues Mol Biol. 2024. PMID: 38248353 Free PMC article.
-
Identification of Allosteric Modulators of Metabotropic Glutamate 7 Receptor Using Proteochemometric Modeling.J Chem Inf Model. 2017 Dec 26;57(12):2976-2985. doi: 10.1021/acs.jcim.7b00338. Epub 2017 Dec 12. J Chem Inf Model. 2017. PMID: 29172488 Free PMC article.
-
Synthesis and Characterization of [18F]JNJ-46356479 as the First 18F-Labeled PET Imaging Ligand for Metabotropic Glutamate Receptor 2.Mol Imaging Biol. 2021 Aug;23(4):527-536. doi: 10.1007/s11307-021-01586-0. Epub 2021 Feb 8. Mol Imaging Biol. 2021. PMID: 33559035 Free PMC article.
References
-
- Ball M, Boyd A, Churchill G, Cuthbert M, Drew M, Fielding M, et al (2012). Isoindolone formation via intramolecular Diels–Alder reaction. Org. Process Res. Dev. 16: 741–747.
-
- Cheng Y, Prusoff WH (1973). Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22: 3099–3108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

