Short linear motifs - ex nihilo evolution of protein regulation
- PMID: 26589632
- PMCID: PMC4654906
- DOI: 10.1186/s12964-015-0120-z
Short linear motifs - ex nihilo evolution of protein regulation
Abstract
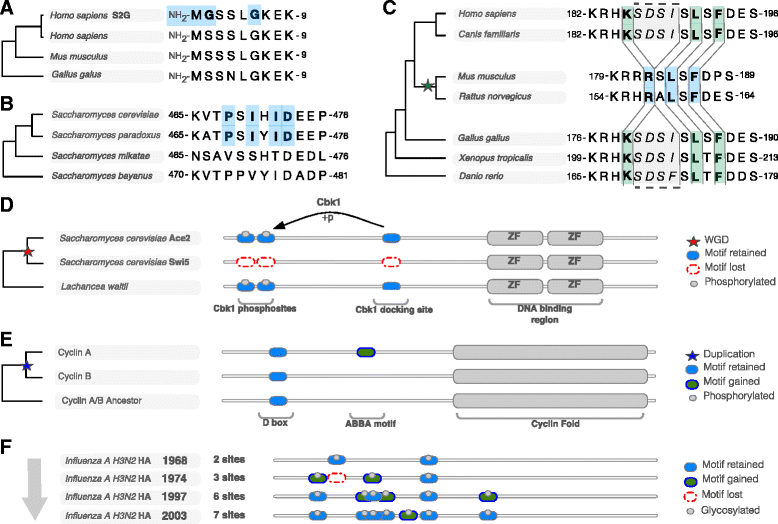
Short sequence motifs are ubiquitous across the three major types of biomolecules: hundreds of classes and thousands of instances of DNA regulatory elements, RNA motifs and protein short linear motifs (SLiMs) have been characterised. The increase in complexity of transcriptional, post-transcriptional and post-translational regulation in higher Eukaryotes has coincided with a significant expansion of motif use. But how did the eukaryotic cell acquire such a vast repertoire of motifs? In this review, we curate the available literature on protein motif evolution and discuss the evidence that suggests SLiMs can be acquired by mutations, insertions and deletions in disordered regions. We propose a mechanism of ex nihilo SLiM evolution - the evolution of a novel SLiM from "nothing" - adding a functional module to a previously non-functional region of protein sequence. In our model, hundreds of motif-binding domains in higher eukaryotic proteins connect simple motif specificities with useful functions to create a large functional motif space. Accessible peptides that match the specificity of these motif-binding domains are continuously created and destroyed by mutations in rapidly evolving disordered regions, creating a dynamic supply of new interactions that may have advantageous phenotypic novelty. This provides a reservoir of diversity to modify existing interaction networks. Evolutionary pressures will act on these motifs to retain beneficial instances. However, most will be lost on an evolutionary timescale as negative selection and genetic drift act on deleterious and neutral motifs respectively. In light of the parallels between the presented model and the evolution of motifs in the regulatory segments of genes and (pre-)mRNAs, we suggest our understanding of regulatory networks would benefit from the creation of a shared model describing the evolution of transcriptional, post-transcriptional and post-translational regulation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

