Torsins: not your typical AAA+ ATPases
- PMID: 26592310
- PMCID: PMC4872298
- DOI: 10.3109/10409238.2015.1091804
Torsins: not your typical AAA+ ATPases
Abstract
Torsin ATPases (Torsins) belong to the widespread AAA+ (ATPases associated with a variety of cellular activities) family of ATPases, which share structural similarity but have diverse cellular functions. Torsins are outliers in this family because they lack many characteristics of typical AAA+ proteins, and they are the only members of the AAA+ family located in the endoplasmic reticulum and contiguous perinuclear space. While it is clear that Torsins have essential roles in many, if not all metazoans, their precise cellular functions remain elusive. Studying Torsins has significant medical relevance since mutations in Torsins or Torsin-associated proteins result in a variety of congenital human disorders, the most frequent of which is early-onset torsion (DYT1) dystonia, a severe movement disorder. A better understanding of the Torsin system is needed to define the molecular etiology of these diseases, potentially enabling corrective therapy. Here, we provide a comprehensive overview of the Torsin system in metazoans, discuss functional clues obtained from various model systems and organisms and provide a phylogenetic and structural analysis of Torsins and their regulatory cofactors in relation to disease-causative mutations. Moreover, we review recent data that have led to a dramatically improved understanding of these machines at a molecular level, providing a foundation for investigating the molecular defects underlying the associated movement disorders. Lastly, we discuss our ideas on how recent progress may be utilized to inform future studies aimed at determining the cellular role(s) of these atypical molecular machines and their implications for dystonia treatment options.
Keywords: Early-onset torsion dystonia; LAP1; LULL1; endoplasmic reticulum; nuclear envelope; torsinA.
Conflict of interest statement
The authors are funded by NIH (DP2 OD008624-01).
The authors report no declarations of interest.
Figures
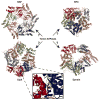

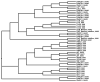

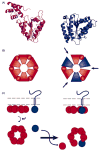

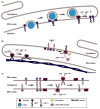

References
-
- BASHAM SE, ROSE LS. Mutations in ooc-5 and ooc-3 disrupt oocyte formation and the reestablishment of asymmetric PAR protein localization in two-cell Caenorhabditis elegans embryos. Dev Biol. 1999;215:253–63. - PubMed
-
- BASHAM SE, ROSE LS. The Caenorhabditis elegans polarity gene ooc-5 encodes a Torsin-related protein of the AAA ATPase superfamily. Development. 2001;128:4645–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
