NDEx, the Network Data Exchange
- PMID: 26594663
- PMCID: PMC4649937
- DOI: 10.1016/j.cels.2015.10.001
NDEx, the Network Data Exchange
Abstract
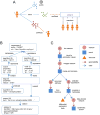
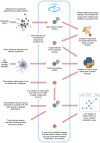
Networks are a powerful and flexible methodology for expressing biological knowledge for computation and communication. Network-encoded information can include systematic screens for molecular interactions, biological relationships curated from literature, and outputs from analysis of Big Data. NDEx, the Network Data Exchange (www.ndexbio.org), is an online commons where scientists can upload, share, and publicly distribute networks. Networks in NDEx receive globally unique accession IDs and can be stored for private use, shared in pre-publication collaboration, or released for public access. Standard and novel data formats are accommodated in a flexible storage model. Organizations can use NDEx as a distribution channel for networks they generate or curate. Developers of bioinformatic applications can store and query NDEx networks via a common programmatic interface. NDEx helps expand the role of networks in scientific discourse and facilitates the integration of networks as data in publications. It is a step towards an ecosystem in which networks bearing data, hypotheses, and findings flow easily between scientists.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

