Conserved mRNA-binding proteomes in eukaryotic organisms
- PMID: 26595419
- PMCID: PMC5759928
- DOI: 10.1038/nsmb.3128
Conserved mRNA-binding proteomes in eukaryotic organisms
Abstract
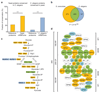
RNA-binding proteins (RBPs) are essential for post-transcriptional regulation of gene expression. Recent high-throughput screens have dramatically increased the number of experimentally identified RBPs; however, comprehensive identification of RBPs within living organisms is elusive. Here we describe the repertoire of 765 and 594 proteins that reproducibly interact with polyadenylated mRNAs in Saccharomyces cerevisiae and Caenorhabditis elegans, respectively. Furthermore, we report the differential association of mRNA-binding proteins (mRPBs) upon induction of apoptosis in C. elegans L4-stage larvae. Strikingly, most proteins composing mRBPomes, including components of early metabolic pathways and the proteasome, are evolutionarily conserved between yeast and C. elegans. We speculate, on the basis of our evidence that glycolytic enzymes bind distinct glycolytic mRNAs, that enzyme-mRNA interactions relate to an ancient mechanism for post-transcriptional coordination of metabolic pathways that perhaps was established during the transition from the early 'RNA world' to the 'protein world'.
Figures


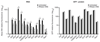

References
-
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

