Update of the human and mouse Fanconi anemia genes
- PMID: 26596371
- PMCID: PMC4657327
- DOI: 10.1186/s40246-015-0054-y
Update of the human and mouse Fanconi anemia genes
Abstract
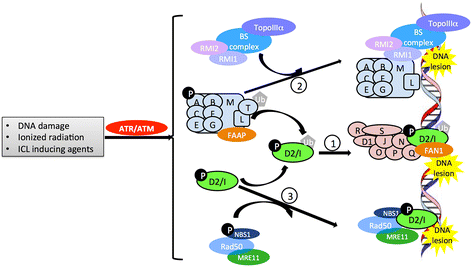
Fanconi anemia (FA) is a recessively inherited disease manifesting developmental abnormalities, bone marrow failure, and increased risk of malignancies. Whereas FA has been studied for nearly 90 years, only in the last 20 years have increasing numbers of genes been implicated in the pathogenesis associated with this genetic disease. To date, 19 genes have been identified that encode Fanconi anemia complementation group proteins, all of which are named or aliased, using the root symbol "FANC." Fanconi anemia subtype (FANC) proteins function in a common DNA repair pathway called "the FA pathway," which is essential for maintaining genomic integrity. The various FANC mutant proteins contribute to distinct steps associated with FA pathogenesis. Herein, we provide a review update of the 19 human FANC and their mouse orthologs, an evolutionary perspective on the FANC genes, and the functional significance of the FA DNA repair pathway in association with clinical disorders. This is an example of a set of genes--known to exist in vertebrates, invertebrates, plants, and yeast--that are grouped together on the basis of shared biochemical and physiological functions, rather than evolutionary phylogeny, and have been named on this basis by the HUGO Gene Nomenclature Committee (HGNC).
Figures

Comment in
-
Letter to the editor for "Update of the human and mouse Fanconi anemia genes".Hum Genomics. 2016 Jul 4;10(1):25. doi: 10.1186/s40246-016-0081-3. Hum Genomics. 2016. PMID: 27377885 Free PMC article. No abstract available.
References
-
- Alter BP, and Kupfer G (1993–2015) Fanconi Anemia. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle. Available From http://www.ncbi.nlm.nih.gov/books/NBK1401/ Access date February 7, 2013.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

