Down-regulation of tomato PHYTOL KINASE strongly impairs tocopherol biosynthesis and affects prenyllipid metabolism in an organ-specific manner
- PMID: 26596763
- PMCID: PMC4737080
- DOI: 10.1093/jxb/erv504
Down-regulation of tomato PHYTOL KINASE strongly impairs tocopherol biosynthesis and affects prenyllipid metabolism in an organ-specific manner
Abstract
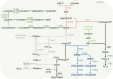
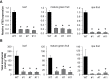
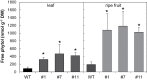
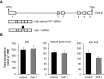
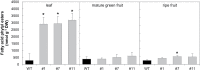
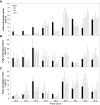
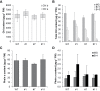
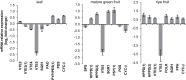
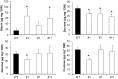
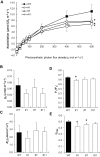
Tocopherol, a compound with vitamin E (VTE) activity, is a conserved constituent of the plastidial antioxidant network in photosynthetic organisms. The synthesis of tocopherol involves the condensation of an aromatic head group with an isoprenoid prenyl side chain. The latter, phytyl diphosphate, can be derived from chlorophyll phytol tail recycling, which depends on phytol kinase (VTE5) activity. How plants co-ordinate isoprenoid precursor distribution for supplying biosynthesis of tocopherol and other prenyllipids in different organs is poorly understood. Here, Solanum lycopersicum plants impaired in the expression of two VTE5-like genes identified by phylogenetic analyses, named SlVTE5 and SlFOLK, were characterized. Our data show that while SlFOLK does not affect tocopherol content, the production of this metabolite is >80% dependent on SlVTE5 in tomato, in both leaves and fruits. VTE5 deficiency greatly impacted lipid metabolism, including prenylquinones, carotenoids, and fatty acid phytyl esters. However, the prenyllipid profile greatly differed between source and sink organs, revealing organ-specific metabolic adjustments in tomato. Additionally, VTE5-deficient plants displayed starch accumulation and lower CO2 assimilation in leaves associated with mild yield penalty. Taken together, our results provide valuable insights into the distinct regulation of isoprenoid metabolism in leaves and fruits and also expose the interaction between lipid and carbon metabolism, which results in carbohydrate export blockage in the VTE5-deficient plants, affecting tomato fruit quality.
Keywords: Carotenoids; Solanum lycopersicum; chlorophyll; phytol; phytol kinase; prenyllipids; tocopherol; tomato; vitamin E..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures
References
-
- Adams WW, Muller O, Cohu CM, Demmig-Adams B. 2013. May photoinhibition be a consequence, rather than a cause, of limited plant productivity? Photosynthesis Research 117, 31–44. - PubMed
-
- Almeida J, Asís R, Molineri VN, Sestari I, Lira BS, Carrari F, Peres LE, Rossi M. 2015. Fruits from ripening impaired, chlorophyll degraded and jasmonate insensitive tomato mutants have altered tocopherol content and composition. Phytochemistry 111, 72–83. - PubMed
-
- Amaral LIV, Gaspar M, Costa PF, Aidar MPM, Buckeridge MS. 2008. Novo método enzimático rápido e sensível de extração e dosagem de amido em materiais vegetais. Hoehnea 34, 425–431.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

