Reading between the Lines: "ADD"-ing Histone and DNA Methylation Marks toward a New Epigenetic "Sum"
- PMID: 26596909
- PMCID: PMC4798892
- DOI: 10.1021/acschembio.5b00830
Reading between the Lines: "ADD"-ing Histone and DNA Methylation Marks toward a New Epigenetic "Sum"
Erratum in
-
Reading between the Lines: "ADD"-ing Histone and DNA Methylation Marks toward a New Epigenetic "Sum".ACS Chem Biol. 2018 Apr 20;13(4):1103. doi: 10.1021/acschembio.8b00162. Epub 2018 Mar 16. ACS Chem Biol. 2018. PMID: 29547264 No abstract available.
Abstract
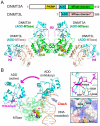
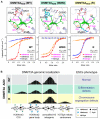
Covalent modifications of both DNA and histones act in concert to define the landscape of our epigenome. In this review, we explore the interconnections between histone and DNA modifications by focusing on a conserved chromatin-binding regulatory domain, the ATRX-DNMT3-DNMT3L (ADD) domain. New studies show that the ADD domain is capable of sensing, and therefore integrating, the status of multiple histone modifications. This in turn dictates the in vivo localization or allosteric regulation of the full-length ADD-containing protein and its ability to function in downstream chromatin remodeling events. Strategies to re-engineer the ADD "reader pocket" in the de novo DNA methyltransferase DNMT3A such that it redirects this "writer" to new genomic loci proved useful in understanding important biological downstream consequences of mis-targeting of DNA methylation via altered reading of histone marks. Combined with genome-editing tools, this approach stands as a poof-of-principle and will be broadly applicable to the elucidation of epigenetic networks that have been altered by "reader" mutations, either artificially or as naturally occurs in some human diseases.
Figures


References
-
- Allis CD, Caparros M-L, Jenuwein T, Reinberg D. Epigenetics. 2nd ed. Chapter 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2015. Overview and Concepts.
-
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. - PubMed
-
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. - PubMed
-
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. - PubMed
-
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

