Role of Mitochondrial RNA Polymerase in the Toxicity of Nucleotide Inhibitors of Hepatitis C Virus
- PMID: 26596942
- PMCID: PMC4750701
- DOI: 10.1128/AAC.01922-15
Role of Mitochondrial RNA Polymerase in the Toxicity of Nucleotide Inhibitors of Hepatitis C Virus
Abstract
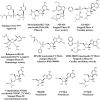
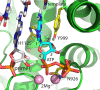
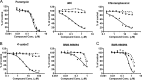
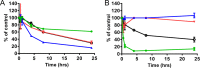
Toxicity has emerged during the clinical development of many but not all nucleotide inhibitors (NI) of hepatitis C virus (HCV). To better understand the mechanism for adverse events, clinically relevant HCV NI were characterized in biochemical and cellular assays, including assays of decreased viability in multiple cell lines and primary cells, interaction with human DNA and RNA polymerases, and inhibition of mitochondrial protein synthesis and respiration. NI that were incorporated by the mitochondrial RNA polymerase (PolRMT) inhibited mitochondrial protein synthesis and showed a corresponding decrease in mitochondrial oxygen consumption in cells. The nucleoside released by the prodrug balapiravir (R1626), 4'-azido cytidine, was a highly selective inhibitor of mitochondrial RNA transcription. The nucleotide prodrug of 2'-C-methyl guanosine, BMS-986094, showed a primary effect on mitochondrial function at submicromolar concentrations, followed by general cytotoxicity. In contrast, NI containing multiple ribose modifications, including the active forms of mericitabine and sofosbuvir, were poor substrates for PolRMT and did not show mitochondrial toxicity in cells. In general, these studies identified the prostate cell line PC-3 as more than an order of magnitude more sensitive to mitochondrial toxicity than the commonly used HepG2 cells. In conclusion, analogous to the role of mitochondrial DNA polymerase gamma in toxicity caused by some 2'-deoxynucleotide analogs, there is an association between HCV NI that interact with PolRMT and the observation of adverse events. More broadly applied, the sensitive methods for detecting mitochondrial toxicity described here may help in the identification of mitochondrial toxicity prior to clinical testing.
Copyright © 2016 Feng et al.
Figures

References
-
- Le Pogam S, Seshaadri A, Kosaka A, Chiu S, Kang H, Hu S, Rajyaguru S, Symons J, Cammack N, Najera I. 2008. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J Antimicrob Chemother 61:1205–1216. doi: 10.1093/jac/dkn085. - DOI - PubMed
-
- McCown MF, Rajyaguru S, Le Pogam S, Ali S, Jiang WR, Kang H, Symons J, Cammack N, Najera I. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob Agents Chemother 52:1604–1612. doi: 10.1128/AAC.01317-07. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

