Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid
- PMID: 26597788
- PMCID: PMC4665766
- DOI: 10.1038/srep17185
Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid
Abstract
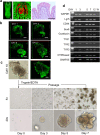
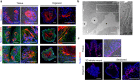
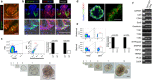
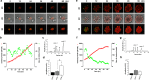
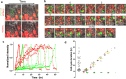
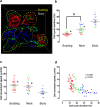
Leucine-rich repeat-containing G-protein coupled receptor 5-expressing (Lgr5(+)) cells have been identified as stem/progenitor cells in the circumvallate papillae, and single cultured Lgr5(+) cells give rise to taste cells. Here we use circumvallate papilla tissue to establish a three-dimensional culture system (taste bud organoids) that develops phenotypic characteristics similar to native tissue, including a multilayered epithelium containing stem/progenitor in the outer layers and taste cells in the inner layers. Furthermore, characterization of the cell cycle of the taste bud progenitor niche reveals striking dynamics of taste bud development and regeneration. Using this taste bud organoid culture system and FUCCI2 transgenic mice, we identify the stem/progenitor cells have at least 5 distinct cell cycle populations by tracking within 24-hour synchronized oscillations of proliferation. Additionally, we demonstrate that stem/progenitor cells have motility to form taste bud organoids. Taste bud organoids provides a system for elucidating mechanisms of taste signaling, disease modeling, and taste tissue regeneration.
Figures


References
-
- Ozdener H. et al. Characterization and long-term maintenance of rat taste cells in culture. Chem Senses 31, 279–290 (2006). - PubMed
-
- Kishi M., Emori Y., Tsukamoto Y. & Abe K. Primary culture of rat taste bud cells that retain molecular markers for taste buds and permit functional expression of foreign genes. Neuroscience 106, 217–225 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

