Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces
- PMID: 26597961
- PMCID: PMC4688200
- DOI: 10.1007/s00248-015-0711-7
Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces
Abstract
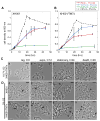
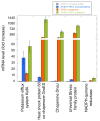
Despite many examples of obligate epibiotic symbiosis (one organism living on the surface of another) in nature, such an interaction has rarely been observed between two bacteria. Here, we further characterize a newly reported interaction between a human oral obligate parasitic bacterium TM7x (cultivated member of Candidatus Saccharimonas formerly Candidate Phylum TM7), and its basibiont Actinomyces odontolyticus species (XH001), providing a model system to study epiparasitic symbiosis in the domain Bacteria. Detailed microscopic studies indicate that both partners display extensive morphological changes during symbiotic growth. XH001 cells manifested as short rods in monoculture, but displayed elongated and hyphal morphology when physically associated with TM7x. Interestingly, these dramatic morphological changes in XH001 were also induced in oxygen-depleted conditions, even in the absence of TM7x. Targeted quantitative real-time PCR (qRT-PCR) analyses revealed that both the physical association with TM7x as well as oxygen depletion triggered up-regulation of key stress response genes in XH001, and in combination, these conditions act in an additive manner. TM7x and XH001 co-exist with relatively uniform cell morphologies under nutrient-replete conditions. However, upon nutrient depletion, TM7x-associated XH001 displayed a variety of cell morphologies, including swollen cell body, clubbed-ends, and even cell lysis, and a large portion of TM7x cells transformed from ultrasmall cocci into elongated cells. Our study demonstrates a highly dynamic interaction between epibiont TM7x and its basibiont XH001 in response to physical association or environmental cues such as oxygen level and nutritional status, as reflected by their morphological and physiological changes during symbiotic growth.
Keywords: Actinomyces; Bacterial interaction; Epibiont; Obligate; Symbiosis; TM7.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
-
- Joseph S. Symbiosis: Mechanisms and Model Systems. Kluwer Academic Publisher; New York, Boston, Dordrecht, London, Moscow: 2002.
-
- Lambina VA, Afinogenova AV, Romaĭ Penabad S, et al. Micavibrio admirandus gen. et sp. nov. Mikrobiologiia. 1982;51:114–117. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

