Enhanced Direct Major Histocompatibility Complex Class I Self-Antigen Presentation Induced by Chlamydia Infection
- PMID: 26597986
- PMCID: PMC4730583
- DOI: 10.1128/IAI.01254-15
Enhanced Direct Major Histocompatibility Complex Class I Self-Antigen Presentation Induced by Chlamydia Infection
Abstract
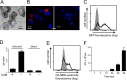
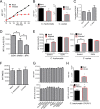
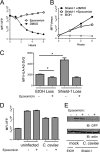
The direct major histocompatibility complex (MHC) class I antigen presentation pathway ensures intracellular peptides are displayed at the cellular surface for recognition of infected or transformed cells by CD8(+) cytotoxic T lymphocytes. Chlamydia spp. are obligate intracellular bacteria and, as such, should be targeted by CD8(+) T cells. It is likely that Chlamydia spp. have evolved mechanisms to avoid the CD8(+) killer T cell responses by interfering with MHC class I antigen presentation. Using a model system of self-peptide presentation which allows for posttranslational control of the model protein's stability, we tested the ability of various Chlamydia species to alter direct MHC class I antigen presentation. Infection of the JY lymphoblastoid cell line limited the accumulation of a model host protein and increased presentation of the model-protein-derived peptides. Enhanced self-peptide presentation was detected only when presentation was restricted to defective ribosomal products, or DRiPs, and total MHC class I levels remained unaltered. Skewed antigen presentation was dependent on a bacterial synthesized component, as evidenced by reversal of the observed phenotype upon preventing bacterial transcription, translation, and the inhibition of bacterial lipooligosaccharide synthesis. These data suggest that Chlamydia spp. have evolved to alter the host antigen presentation machinery to favor presentation of defective and rapidly degraded forms of self-antigen, possibly as a mechanism to diminish the presentation of peptides derived from bacterial proteins.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Chlamydia trachomatis Infection Impairs MHC-I Intracellular Trafficking and Antigen Cross-Presentation by Dendritic Cells.Front Immunol. 2021 Apr 15;12:662096. doi: 10.3389/fimmu.2021.662096. eCollection 2021. Front Immunol. 2021. PMID: 33936099 Free PMC article.
-
Discordance in the Epithelial Cell-Dendritic Cell Major Histocompatibility Complex Class II Immunoproteome: Implications for Chlamydia Vaccine Development.J Infect Dis. 2020 Feb 18;221(5):841-850. doi: 10.1093/infdis/jiz522. J Infect Dis. 2020. PMID: 31599954 Free PMC article.
-
Functional characterization of class Ia- and non-class Ia-restricted Chlamydia-reactive CD8+ T cell responses in humans.J Immunol. 2003 Oct 15;171(8):4278-86. doi: 10.4049/jimmunol.171.8.4278. J Immunol. 2003. PMID: 14530352
-
Involvement of autophagy in MHC class I antigen presentation.Scand J Immunol. 2020 Nov;92(5):e12978. doi: 10.1111/sji.12978. Epub 2020 Oct 19. Scand J Immunol. 2020. PMID: 32969499 Free PMC article. Review.
-
Role of autophagy in MHC class I-restricted antigen presentation.Mol Immunol. 2019 Sep;113:2-5. doi: 10.1016/j.molimm.2017.10.021. Epub 2017 Nov 8. Mol Immunol. 2019. PMID: 29126597 Free PMC article. Review.
Cited by
-
Chlamydial Infection From Outside to Inside.Front Microbiol. 2019 Oct 9;10:2329. doi: 10.3389/fmicb.2019.02329. eCollection 2019. Front Microbiol. 2019. PMID: 31649655 Free PMC article. Review.
-
Human Fallopian Tube Epithelial Cell Culture Model To Study Host Responses to Chlamydia trachomatis Infection.Infect Immun. 2020 Aug 19;88(9):e00105-20. doi: 10.1128/IAI.00105-20. Print 2020 Aug 19. Infect Immun. 2020. PMID: 32601108 Free PMC article.
-
Inhibition of the Deubiquitinase Usp14 Diminishes Direct MHC Class I Antigen Presentation.J Immunol. 2018 Feb 1;200(3):928-936. doi: 10.4049/jimmunol.1700273. Epub 2017 Dec 27. J Immunol. 2018. PMID: 29282303 Free PMC article.
-
Chlamydia Lipooligosaccharide Has Varied Direct and Indirect Roles in Evading both Innate and Adaptive Host Immune Responses.Infect Immun. 2020 Jul 21;88(8):e00198-20. doi: 10.1128/IAI.00198-20. Print 2020 Jul 21. Infect Immun. 2020. PMID: 32423914 Free PMC article.
-
Chlamydia spp. development is differentially altered by treatment with the LpxC inhibitor LPC-011.BMC Microbiol. 2017 Apr 24;17(1):98. doi: 10.1186/s12866-017-0992-8. BMC Microbiol. 2017. PMID: 28438125 Free PMC article.
References
-
- Holland MJ, Faal N, Sarr I, Joof H, Laye M, Cameron E, Pemberton-Pigott F, Dockrell HM, Bailey RL, Mabey DCW. 2006. The frequency of Chlamydia trachomatis major outer membrane protein-specific CD8+ T lymphocytes in active trachoma is associated with current ocular infection. Infect Immun 74:1565–1572. doi:10.1128/IAI.74.3.1565-1572.2006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

