The Chromosome-Encoded Hypothetical Protein TC0668 Is an Upper Genital Tract Pathogenicity Factor of Chlamydia muridarum
- PMID: 26597987
- PMCID: PMC4730586
- DOI: 10.1128/IAI.01171-15
The Chromosome-Encoded Hypothetical Protein TC0668 Is an Upper Genital Tract Pathogenicity Factor of Chlamydia muridarum
Abstract
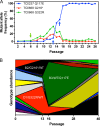
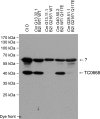
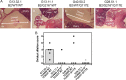
We previously associated a missense mutation of the tc0668 gene of serial in vitro-passaged Chlamydia muridarum, a murine model of human urogenital C. trachomatis, with severely attenuated disease development in the upper genital tract of female mice. Since these mutants also contained a TC0237 Q117E missense mutation that enhances their in vitro infectivity, an effort was made here to isolate and characterize a tc0668 single mutant to determine its individual contribution to urogenital pathogenicity. Detailed genetic analysis of C. muridarum passages revealed a truncated variant with a G216* nonsense mutation of the 408-amino-acid TC0668 protein that does not produce a detectable product. Intracellular growth and infectivity of C. muridarum in vitro remain unaffected in the absence of TC0668. Intravaginal inoculation of the TC0668 null mutant into C3H/HeJ mice results in a typical course of lower genital tract infection but, unlike a pathogenic isogenic control, is unable to elicit significant chronic inflammation of the oviduct and fails to induce hydrosalpinx. Thus, TC0668 is demonstrated as an important chromosome-encoded urogenital pathogenicity factor of C. muridarum and the first with these characteristics to be discovered for a Chlamydia pathogen.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
In vitro passage selects for Chlamydia muridarum with enhanced infectivity in cultured cells but attenuated pathogenicity in mouse upper genital tract.Infect Immun. 2015 May;83(5):1881-92. doi: 10.1128/IAI.03158-14. Epub 2015 Feb 23. Infect Immun. 2015. PMID: 25712926 Free PMC article.
-
Chlamydia muridarum with Mutations in Chromosomal Genes tc0237 and/or tc0668 Is Deficient in Colonizing the Mouse Gastrointestinal Tract.Infect Immun. 2017 Jul 19;85(8):e00321-17. doi: 10.1128/IAI.00321-17. Print 2017 Aug. Infect Immun. 2017. PMID: 28584162 Free PMC article.
-
A primary study on genes with selected mutations by in vitro passage of Chlamydia muridarum strains.Pathog Dis. 2019 Apr 1;77(3):ftz017. doi: 10.1093/femspd/ftz017. Pathog Dis. 2019. PMID: 31197357 Free PMC article.
-
Chlamydia Spreading from the Genital Tract to the Gastrointestinal Tract - A Two-Hit Hypothesis.Trends Microbiol. 2018 Jul;26(7):611-623. doi: 10.1016/j.tim.2017.12.002. Epub 2017 Dec 27. Trends Microbiol. 2018. PMID: 29289422 Free PMC article. Review.
-
Chlamydia overcomes multiple gastrointestinal barriers to achieve long-lasting colonization.Trends Microbiol. 2021 Nov;29(11):1004-1012. doi: 10.1016/j.tim.2021.03.011. Epub 2021 Apr 14. Trends Microbiol. 2021. PMID: 33865675 Free PMC article. Review.
Cited by
-
Chlamydia Spreads to the Large Intestine Lumen via Multiple Pathways.Infect Immun. 2021 Sep 16;89(10):e0025421. doi: 10.1128/IAI.00254-21. Epub 2021 Jul 19. Infect Immun. 2021. PMID: 34280037 Free PMC article.
-
Development of Transposon Mutagenesis for Chlamydia muridarum.J Bacteriol. 2019 Nov 5;201(23):e00366-19. doi: 10.1128/JB.00366-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501283 Free PMC article.
-
Evasion of Innate Lymphoid Cell-Regulated Gamma Interferon Responses by Chlamydia muridarum To Achieve Long-Lasting Colonization in Mouse Colon.Infect Immun. 2020 Feb 20;88(3):e00798-19. doi: 10.1128/IAI.00798-19. Print 2020 Feb 20. Infect Immun. 2020. PMID: 31818961 Free PMC article.
-
Chlamydia muridarum Induces Pathology in the Female Upper Genital Tract via Distinct Mechanisms.Infect Immun. 2019 Jul 23;87(8):e00145-19. doi: 10.1128/IAI.00145-19. Print 2019 Aug. Infect Immun. 2019. PMID: 31085708 Free PMC article.
-
The emerging role of ASC in dendritic cell metabolism during Chlamydia infection.PLoS One. 2017 Dec 7;12(12):e0188643. doi: 10.1371/journal.pone.0188643. eCollection 2017. PLoS One. 2017. PMID: 29216217 Free PMC article.
References
-
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi:10.1093/nar/28.6.1397. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

