The role, mechanism and potentially novel biomarker of microRNA-17-92 cluster in macrosomia
- PMID: 26598317
- PMCID: PMC4657041
- DOI: 10.1038/srep17212
The role, mechanism and potentially novel biomarker of microRNA-17-92 cluster in macrosomia
Abstract
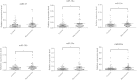
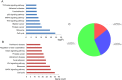
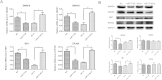
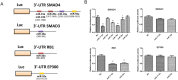
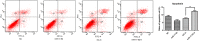
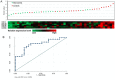
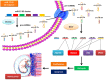
Macrosomia is one of the most common perinatal complications of pregnancy and has life-long health implications for the infant. microRNAs (miRNAs) have been identified to regulate placental development, yet the role of miRNAs in macrosomia remains poorly understood. Here we investigated the role of miR-17-92 cluster in macrosomia. The expression levels of five miRNAs in miR-17-92 cluster were significantly elevated in placentas of macrosomia, which may due to the up-regulation of miRNA-processing enzyme Drosha and Dicer. Cell cycle pathway was identified to be the most relevant pathways regulated by miR-17-92 cluster miRNAs. Importantly, miR-17-92 cluster increased proliferation, attenuated cell apoptosis and accelerated cells entering S phase by targeting SMAD4 and RB1 in HTR8/SVneo cells. Furthermore, we found that expression of miR-17-92 cluster in serum had a high diagnostic sensitivity and specificity for macrosomia (AUC: 80.53%; sensitivity: 82.61%; specificity: 69.57%). Our results suggested that miR-17-92 cluster contribute to macrosomia development by targeting regulators of cell cycle pathway. Our findings not only provide a novel insight into the molecular mechanisms of macrosomia, but also the clinical value of miR-17-92 cluster as a predictive biomarker for macrosomia.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Wei J. N. et al. Low birth weight and high birth weight infants are both at an increased risk to have type 2 diabetes among schoolchildren in taiwan. Diabetes care 26, 343–348 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

