Functional Analysis of Bacteriophage Immunity through a Type I-E CRISPR-Cas System in Vibrio cholerae and Its Application in Bacteriophage Genome Engineering
- PMID: 26598368
- PMCID: PMC4719448
- DOI: 10.1128/JB.00747-15
Functional Analysis of Bacteriophage Immunity through a Type I-E CRISPR-Cas System in Vibrio cholerae and Its Application in Bacteriophage Genome Engineering
Abstract
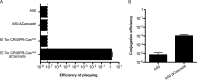
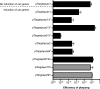
The classical and El Tor biotypes of Vibrio cholerae serogroup O1, the etiological agent of cholera, are responsible for the sixth and seventh (current) pandemics, respectively. A genomic island (GI), GI-24, previously identified in a classical biotype strain of V. cholerae, is predicted to encode clustered regularly interspaced short palindromic repeat (CRISPR)-associated proteins (Cas proteins); however, experimental evidence in support of CRISPR activity in V. cholerae has not been documented. Here, we show that CRISPR-Cas is ubiquitous in strains of the classical biotype but excluded from strains of the El Tor biotype. We also provide in silico evidence to suggest that CRISPR-Cas actively contributes to phage resistance in classical strains. We demonstrate that transfer of GI-24 to V. cholerae El Tor via natural transformation enables CRISPR-Cas-mediated resistance to bacteriophage CP-T1 under laboratory conditions. To elucidate the sequence requirements of this type I-E CRISPR-Cas system, we engineered a plasmid-based system allowing the directed targeting of a region of interest. Through screening for phage mutants that escape CRISPR-Cas-mediated resistance, we show that CRISPR targets must be accompanied by a 3' TT protospacer-adjacent motif (PAM) for efficient interference. Finally, we demonstrate that efficient editing of V. cholerae lytic phage genomes can be performed by simultaneously introducing an editing template that allows homologous recombination and escape from CRISPR-Cas targeting.
Importance: Cholera, caused by the facultative pathogen Vibrio cholerae, remains a serious public health threat. Clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins (CRISPR-Cas) provide prokaryotes with sequence-specific protection from invading nucleic acids, including bacteriophages. In this work, we show that one genomic feature differentiating sixth pandemic (classical biotype) strains from seventh pandemic (El Tor biotype) strains is the presence of a CRISPR-Cas system in the classical biotype. We demonstrate that the CRISPR-Cas system from a classical biotype strain can be transferred to a V. cholerae El Tor biotype strain and that it is functional in providing resistance to phage infection. Finally, we show that this CRISPR-Cas system can be used as an efficient tool for the editing of V. cholerae lytic phage genomes.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi:10.1056/NEJMoa1012928. - DOI - PMC - PubMed
-
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JLN, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi:10.1038/nature10392. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

