A bacterial type III secretion-based protein delivery tool for broad applications in cell biology
- PMID: 26598622
- PMCID: PMC4657163
- DOI: 10.1083/jcb.201502074
A bacterial type III secretion-based protein delivery tool for broad applications in cell biology
Abstract
Methods enabling the delivery of proteins into eukaryotic cells are essential to address protein functions. Here we propose broad applications to cell biology for a protein delivery tool based on bacterial type III secretion (T3S). We show that bacterial, viral, and human proteins, fused to the N-terminal fragment of the Yersinia enterocolitica T3S substrate YopE, are effectively delivered into target cells in a fast and controllable manner via the injectisome of extracellular bacteria. This method enables functional interaction studies by the simultaneous injection of multiple proteins and allows the targeting of proteins to different subcellular locations by use of nanobody-fusion proteins. After delivery, proteins can be freed from the YopE fragment by a T3S-translocated viral protease or fusion to ubiquitin and cleavage by endogenous ubiquitin proteases. Finally, we show that this delivery tool is suitable to inject proteins in living animals and combine it with phosphoproteomics to characterize the systems-level impact of proapoptotic human truncated BID on the cellular network.
© 2015 Ittig et al.
Figures

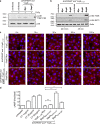
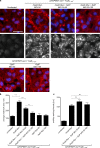




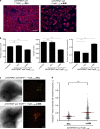
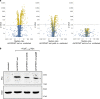

References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

