High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein
- PMID: 26598693
- PMCID: PMC4679053
- DOI: 10.1073/pnas.1513803112
High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein
Abstract
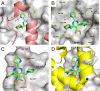
The hepatitis B virus (HBV) core protein is essential for HBV replication and an important target for antiviral drug discovery. We report the first, to our knowledge, high-resolution crystal structure of an antiviral compound bound to the HBV core protein. The compound NVR-010-001-E2 can induce assembly of the HBV core wild-type and Y132A mutant proteins and thermostabilize the proteins with a Tm increase of more than 10 °C. NVR-010-001-E2 binds at the dimer-dimer interface of the core proteins, forms a new interaction surface promoting protein-protein interaction, induces protein assembly, and increases stability. The impact of naturally occurring core protein mutations on antiviral activity correlates with NVR-010-001-E2 binding interactions determined by crystallography. The crystal structure provides understanding of a drug efficacy mechanism related to the induction and stabilization of protein-protein interactions and enables structure-guided design to improve antiviral potency and drug-like properties.
Keywords: HBV inhibitor; HBV treatment; capsid; core; protein–protein interaction.
Conflict of interest statement
Conflict of interest statement: The authors are employees of Novira Therapeutics (K.K., A.M.L., R.V., S.R., C.E., G.H., and O.A.F.), Beryllium (C.L., R.B., K.A., and J.A.), and Vista Informatics Corporation (G.L.).
Figures


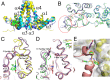
References
-
- Cowie BC, Carville KS, MacLachlan JH. Mortality due to viral hepatitis in the Global Burden of Disease Study 2010: New evidence of an urgent global public health priority demanding action. Antivir Ther. 2013;18(8):953–954. - PubMed
-
- Klumpp K, Crépin T. Capsid proteins of enveloped viruses as antiviral drug targets. Curr Opin Virol. 2014;5:63–71. - PubMed
-
- Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol II. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 2185–2221.
-
- Dryden KA, et al. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell. 2006;22(6):843–850. - PubMed
-
- Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3(6):771–780. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

