A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases
- PMID: 26598975
- PMCID: PMC4798913
- DOI: 10.1021/acschembio.5b00839
A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases
Abstract
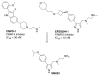
Protein arginine methyltransferases (PRMTs) play a crucial role in a variety of biological processes. Overexpression of PRMTs has been implicated in various human diseases including cancer. Consequently, selective small-molecule inhibitors of PRMTs have been pursued by both academia and the pharmaceutical industry as chemical tools for testing biological and therapeutic hypotheses. PRMTs are divided into three categories: type I PRMTs which catalyze mono- and asymmetric dimethylation of arginine residues, type II PRMTs which catalyze mono- and symmetric dimethylation of arginine residues, and type III PRMT which catalyzes only monomethylation of arginine residues. Here, we report the discovery of a potent, selective, and cell-active inhibitor of human type I PRMTs, MS023, and characterization of this inhibitor in a battery of biochemical, biophysical, and cellular assays. MS023 displayed high potency for type I PRMTs including PRMT1, -3, -4, -6, and -8 but was completely inactive against type II and type III PRMTs, protein lysine methyltransferases and DNA methyltransferases. A crystal structure of PRMT6 in complex with MS023 revealed that MS023 binds the substrate binding site. MS023 potently decreased cellular levels of histone arginine asymmetric dimethylation. It also reduced global levels of arginine asymmetric dimethylation and concurrently increased levels of arginine monomethylation and symmetric dimethylation in cells. We also developed MS094, a close analog of MS023, which was inactive in biochemical and cellular assays, as a negative control for chemical biology studies. MS023 and MS094 are useful chemical tools for investigating the role of type I PRMTs in health and disease.
Figures
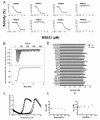
 ) 200 μM MS023 (ΔTm = 20 °C) and (□) 100 μM SAM plus 200 μM MS023 (ΔTm = 22 °C). The inflection point of each transition curve is considered melting temperature (Tm) and the increase in Tm is an indication of binding. (D) The selectivity of MS023 was determined against a panel of 25 PKMTs and DNMTs and 3 histone lysine demethylases at two compound concentrations of 1 μM and 10 μM. No change was observed in the IC50 values when determined under different peptide (E) and SAM (F) concentrations for PRMT6 indicating a noncompetitive pattern of inhibition.
) 200 μM MS023 (ΔTm = 20 °C) and (□) 100 μM SAM plus 200 μM MS023 (ΔTm = 22 °C). The inflection point of each transition curve is considered melting temperature (Tm) and the increase in Tm is an indication of binding. (D) The selectivity of MS023 was determined against a panel of 25 PKMTs and DNMTs and 3 histone lysine demethylases at two compound concentrations of 1 μM and 10 μM. No change was observed in the IC50 values when determined under different peptide (E) and SAM (F) concentrations for PRMT6 indicating a noncompetitive pattern of inhibition.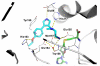


Similar articles
-
Opto-Epigenetic Regulation of Histone Arginine Asymmetric Dimethylation via Type I Protein Arginine Methyltransferase Inhibition.J Med Chem. 2025 Feb 27;68(4):4373-4381. doi: 10.1021/acs.jmedchem.4c02199. Epub 2025 Feb 17. J Med Chem. 2025. PMID: 39961800 Free PMC article.
-
Therapeutic Targeting of RNA Splicing Catalysis through Inhibition of Protein Arginine Methylation.Cancer Cell. 2019 Aug 12;36(2):194-209.e9. doi: 10.1016/j.ccell.2019.07.003. Cancer Cell. 2019. PMID: 31408619 Free PMC article.
-
Discovery of a Potent, Selective, and Cell-Active Dual Inhibitor of Protein Arginine Methyltransferase 4 and Protein Arginine Methyltransferase 6.J Med Chem. 2016 Oct 13;59(19):9124-9139. doi: 10.1021/acs.jmedchem.6b01033. Epub 2016 Sep 15. J Med Chem. 2016. PMID: 27584694 Free PMC article.
-
A patent review of arginine methyltransferase inhibitors (2010-2018).Expert Opin Ther Pat. 2019 Feb;29(2):97-114. doi: 10.1080/13543776.2019.1567711. Expert Opin Ther Pat. 2019. PMID: 30640571 Review.
-
Chemical probes for protein arginine methyltransferases.Methods. 2020 Mar 15;175:30-43. doi: 10.1016/j.ymeth.2019.11.017. Epub 2019 Dec 3. Methods. 2020. PMID: 31809836 Review.
Cited by
-
PRMT1 Is Critical for the Transcriptional Activity and the Stability of the Progesterone Receptor.iScience. 2020 Jun 26;23(6):101236. doi: 10.1016/j.isci.2020.101236. Epub 2020 Jun 4. iScience. 2020. PMID: 32563156 Free PMC article.
-
Type I PRMT Inhibition Protects Against C9ORF72 Arginine-Rich Dipeptide Repeat Toxicity.Front Pharmacol. 2020 Sep 8;11:569661. doi: 10.3389/fphar.2020.569661. eCollection 2020. Front Pharmacol. 2020. PMID: 33013410 Free PMC article.
-
Structure, Activity, and Function of PRMT1.Life (Basel). 2021 Oct 27;11(11):1147. doi: 10.3390/life11111147. Life (Basel). 2021. PMID: 34833023 Free PMC article. Review.
-
PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD+ acute myeloid leukemia.Blood. 2019 Aug 8;134(6):548-560. doi: 10.1182/blood.2019001282. Epub 2019 Jun 19. Blood. 2019. PMID: 31217189 Free PMC article.
-
Pharmacologic modulation of RNA splicing enhances anti-tumor immunity.Cell. 2021 Jul 22;184(15):4032-4047.e31. doi: 10.1016/j.cell.2021.05.038. Epub 2021 Jun 24. Cell. 2021. PMID: 34171309 Free PMC article.
References
-
- Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011;12:629–642. - PubMed
-
- Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of Ng,Ng-Dimethylarginine in Nuclear Protein-Fractions. Biochem. Biophys. Res. Commun. 1977;74:969–976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials

