Computational Modelling of Metastasis Development in Renal Cell Carcinoma
- PMID: 26599078
- PMCID: PMC4658171
- DOI: 10.1371/journal.pcbi.1004626
Computational Modelling of Metastasis Development in Renal Cell Carcinoma
Abstract
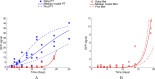
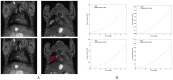
The biology of the metastatic colonization process remains a poorly understood phenomenon. To improve our knowledge of its dynamics, we conducted a modelling study based on multi-modal data from an orthotopic murine experimental system of metastatic renal cell carcinoma. The standard theory of metastatic colonization usually assumes that secondary tumours, once established at a distant site, grow independently from each other and from the primary tumour. Using a mathematical model that translates this assumption into equations, we challenged this theory against our data that included: 1) dynamics of primary tumour cells in the kidney and metastatic cells in the lungs, retrieved by green fluorescent protein tracking, and 2) magnetic resonance images (MRI) informing on the number and size of macroscopic lesions. Critically, when calibrated on the growth of the primary tumour and total metastatic burden, the predicted theoretical size distributions were not in agreement with the MRI observations. Moreover, tumour expansion only based on proliferation was not able to explain the volume increase of the metastatic lesions. These findings strongly suggested rejection of the standard theory, demonstrating that the time development of the size distribution of metastases could not be explained by independent growth of metastatic foci. This led us to investigate the effect of spatial interactions between merging metastatic tumours on the dynamics of the global metastatic burden. We derived a mathematical model of spatial tumour growth, confronted it with experimental data of single metastatic tumour growth, and used it to provide insights on the dynamics of multiple tumours growing in close vicinity. Together, our results have implications for theories of the metastatic process and suggest that global dynamics of metastasis development is dependent on spatial interactions between metastatic lesions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127: 679–695. - PubMed
-
- O’Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Moses M, et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 1994;79: 315–328. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

