Rhinovirus Load Is High despite Preserved Interferon-β Response in Cystic Fibrosis Bronchial Epithelial Cells
- PMID: 26599098
- PMCID: PMC4658124
- DOI: 10.1371/journal.pone.0143129
Rhinovirus Load Is High despite Preserved Interferon-β Response in Cystic Fibrosis Bronchial Epithelial Cells
Abstract
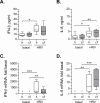
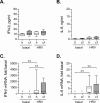
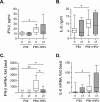
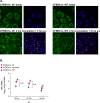
Lung disease in cystic fibrosis (CF) is often exacerbated following acute upper respiratory tract infections caused by the human rhinovirus (HRV). Pathophysiology of these exacerbations is presently unclear and may involve deficient innate antiviral or exaggerated inflammatory responses in CF airway epithelial cells. Furthermore, responses of CF cells to HRV may be adversely affected by pre-exposure to virulence factors of Pseudomonas (P.) aeruginosa, the microorganism that frequently colonizes CF airways. Here we examined production of antiviral cytokine interferon-β and inflammatory chemokine interleukin-8, expression of the interferon-responsive antiviral gene 2'-5'-oligoadenylate synthetase 1 (OAS1), and intracellular virus RNA load in primary CF (delF508 CFTR) and healthy airway epithelial cells following inoculation with HRV16. Parallel cells were exposed to virulence factors of P. aeruginosa prior to and during HRV16 inoculation. CF cells exhibited production of interferon-β and interleukin-8, and expression of OAS1 at levels comparable to those in healthy cells, yet significantly higher HRV16 RNA load during early hours post-inoculation with HRV16. In line with this, HRV16 RNA load was higher in the CFBE41o- dF cell line overexpessing delF508 CFTR, compared with the isogenic control CFBE41o- WT (wild-type CFTR). Pre-exposure to virulence factors of P. aeruginosa did not affect OAS1 expression or HRV16 RNA load, but potentiated interleukin-8 production. In conclusion, CF cells demonstrate elevated HRV RNA load despite preserved interferon-β and OAS1 responses. High HRV load in CF airway epithelial cells appears to be due to deficiencies manifesting early during HRV infection, and may not be related to interferon-β.
Conflict of interest statement
Figures






References
-
- Esposito S, Dacco V, Daleno C, Gambazza S, Montinaro V, Bisogno A, et al. Human rhinovirus infection in children with cystic fibrosis. Jpn J Infect Dis. 2014;67(5):399–401. Epub 2014/09/23. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

