Multi-Parametric MRI and Texture Analysis to Visualize Spatial Histologic Heterogeneity and Tumor Extent in Glioblastoma
- PMID: 26599106
- PMCID: PMC4658019
- DOI: 10.1371/journal.pone.0141506
Multi-Parametric MRI and Texture Analysis to Visualize Spatial Histologic Heterogeneity and Tumor Extent in Glioblastoma
Abstract
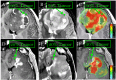
Background: Genetic profiling represents the future of neuro-oncology but suffers from inadequate biopsies in heterogeneous tumors like Glioblastoma (GBM). Contrast-enhanced MRI (CE-MRI) targets enhancing core (ENH) but yields adequate tumor in only ~60% of cases. Further, CE-MRI poorly localizes infiltrative tumor within surrounding non-enhancing parenchyma, or brain-around-tumor (BAT), despite the importance of characterizing this tumor segment, which universally recurs. In this study, we use multiple texture analysis and machine learning (ML) algorithms to analyze multi-parametric MRI, and produce new images indicating tumor-rich targets in GBM.
Methods: We recruited primary GBM patients undergoing image-guided biopsies and acquired pre-operative MRI: CE-MRI, Dynamic-Susceptibility-weighted-Contrast-enhanced-MRI, and Diffusion Tensor Imaging. Following image coregistration and region of interest placement at biopsy locations, we compared MRI metrics and regional texture with histologic diagnoses of high- vs low-tumor content (≥80% vs <80% tumor nuclei) for corresponding samples. In a training set, we used three texture analysis algorithms and three ML methods to identify MRI-texture features that optimized model accuracy to distinguish tumor content. We confirmed model accuracy in a separate validation set.
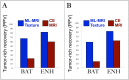
Results: We collected 82 biopsies from 18 GBMs throughout ENH and BAT. The MRI-based model achieved 85% cross-validated accuracy to diagnose high- vs low-tumor in the training set (60 biopsies, 11 patients). The model achieved 81.8% accuracy in the validation set (22 biopsies, 7 patients).
Conclusion: Multi-parametric MRI and texture analysis can help characterize and visualize GBM's spatial histologic heterogeneity to identify regional tumor-rich biopsy targets.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

