What Is the Best NGS Enrichment Method for the Molecular Diagnosis of Monogenic Diabetes and Obesity?
- PMID: 26599467
- PMCID: PMC4657897
- DOI: 10.1371/journal.pone.0143373
What Is the Best NGS Enrichment Method for the Molecular Diagnosis of Monogenic Diabetes and Obesity?
Abstract
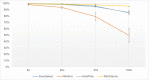
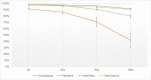
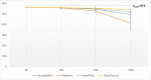
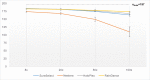
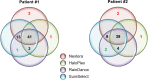
Molecular diagnosis of monogenic diabetes and obesity is of paramount importance for both the patient and society, as it can result in personalized medicine associated with a better life and it eventually saves health care spending. Genetic clinical laboratories are currently switching from Sanger sequencing to next-generation sequencing (NGS) approaches but choosing the optimal protocols is not easy. Here, we compared the sequencing coverage of 43 genes involved in monogenic forms of diabetes and obesity, and variant detection rates, resulting from four enrichment methods based on the sonication of DNA (Agilent SureSelect, RainDance technologies), or using enzymes for DNA fragmentation (Illumina Nextera, Agilent HaloPlex). We analyzed coding exons and untranslated regions of the 43 genes involved in monogenic diabetes and obesity. We found that none of the methods achieves yet full sequencing of the gene targets. Nonetheless, the RainDance, SureSelect and HaloPlex enrichment methods led to the best sequencing coverage of the targets; while the Nextera method resulted in the poorest sequencing coverage. Although the sequencing coverage was high, we unexpectedly found that the HaloPlex method missed 20% of variants detected by the three other methods and Nextera missed 10%. The question of which NGS technique for genetic diagnosis yields the highest diagnosis rate is frequently discussed in the literature and the response is still unclear. Here, we showed that the RainDance enrichment method as well as SureSelect, which are both based on the sonication of DNA, resulted in a good sequencing quality and variant detection, while the use of enzymes to fragment DNA (HaloPlex or Nextera) might not be the best strategy to get an accurate sequencing.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

