Vaginal Microbiome Characterization of Nellore Cattle Using Metagenomic Analysis
- PMID: 26599789
- PMCID: PMC4657983
- DOI: 10.1371/journal.pone.0143294
Vaginal Microbiome Characterization of Nellore Cattle Using Metagenomic Analysis
Abstract
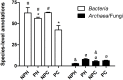
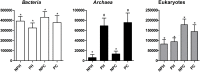
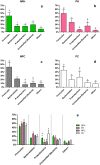
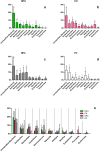
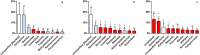
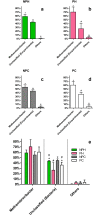
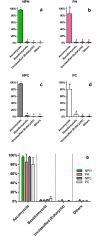
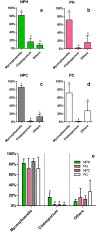
Understanding of microbial communities inhabiting cattle vaginal tract may lead to a better comprehension of bovine physiology and reproductive health being of great economic interest. Up to date, studies involving cattle microbiota are focused on the gastrointestinal tract, and little is known about the vaginal microbiota. This study aimed to investigate the vaginal microbiome in Nellore cattle, heifers and cows, pregnant and non-pregnant, using a culture independent approach. The main bacterial phyla found were Firmicutes (~40-50%), Bacteroidetes (~15-25%) and Proteobacteria (~5-25%), in addition to ~10-20% of non-classified bacteria. 45-55% of the samples were represented by only ten OTUs: Aeribacillus, Bacteroides, Clostridium, Ruminococcus, Rikenella, Alistipes, Bacillus, Eubacterium, Prevotella and non-classified bacteria. Interestingly, microbiota from all 20 animals could be grouped according to the respiratory metabolism of the main OTUs found, creating three groups of vaginal microbiota in cattle. Archaeal samples were dominated by the Methanobrevibacter genus (Euryarchaeota, ~55-70%). Ascomycota was the main fungal phylum (~80-95%) and Mycosphaerella the most abundant genus (~70-85%). Hormonal influence was not clear, but a tendency for the reduction of bacterial and increase of archaeal populations in pregnant animals was observed. Eukaryotes did not vary significantly between pregnant and non-pregnant animals, but tended to be more abundant on cows than on heifers. The present work describes a great microbial variability in the vaginal community among the evaluated animals and groups (heifers and cows, pregnant and non-pregnant), which is significantly different from the findings previously reported using culture dependent methods, pointing out the need for further studies on this issue. The microbiome found also indicates that the vaginal colonization appears to be influenced by the gastrointestinal community.
Conflict of interest statement
Figures

References
-
- United States Department of Agriculture (USDA). Livestock and poultry: World markets and trade. 2014.
-
- Dumonceaux TJ, Schellenberg J, Goleski V, Hill JE, Jaoko W, Kimani J, et al. Multiplex detection of bacteria associated with normal microbiota and with bacterial vaginosis in vaginal swabs by use of oligonucleotide-coupled fluorescent microspheres. J Clin Microbiol. 2009;47: 4067–4077. 10.1128/JCM.00112-09 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

