SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis
- PMID: 26600239
- PMCID: PMC4658017
- DOI: 10.1371/journal.pcbi.1004575
SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis
Abstract
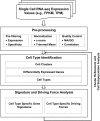
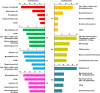
A major challenge in developmental biology is to understand the genetic and cellular processes/programs driving organ formation and differentiation of the diverse cell types that comprise the embryo. While recent studies using single cell transcriptome analysis illustrate the power to measure and understand cellular heterogeneity in complex biological systems, processing large amounts of RNA-seq data from heterogeneous cell populations creates the need for readily accessible tools for the analysis of single-cell RNA-seq (scRNA-seq) profiles. The present study presents a generally applicable analytic pipeline (SINCERA: a computational pipeline for SINgle CEll RNA-seq profiling Analysis) for processing scRNA-seq data from a whole organ or sorted cells. The pipeline supports the analysis for: 1) the distinction and identification of major cell types; 2) the identification of cell type specific gene signatures; and 3) the determination of driving forces of given cell types. We applied this pipeline to the RNA-seq analysis of single cells isolated from embryonic mouse lung at E16.5. Through the pipeline analysis, we distinguished major cell types of fetal mouse lung, including epithelial, endothelial, smooth muscle, pericyte, and fibroblast-like cell types, and identified cell type specific gene signatures, bioprocesses, and key regulators. SINCERA is implemented in R, licensed under the GNU General Public License v3, and freely available from CCHMC PBGE website, https://research.cchmc.org/pbge/sincera.html.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Neildez-Nguyen TM, Parisot A, Vignal C, Rameau P, Stockholm D, Picot J, Allo V, Le Bec C, Laplace C, Paldi A (2008) Epigenetic gene expression noise and phenotypic diversification of clonal cell populations. Differentiation 76: 33–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

