Biodiversity and distribution of polar freshwater DNA viruses
- PMID: 26601189
- PMCID: PMC4640604
- DOI: 10.1126/sciadv.1400127
Biodiversity and distribution of polar freshwater DNA viruses
Abstract
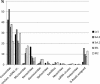
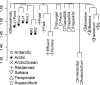
Viruses constitute the most abundant biological entities and a large reservoir of genetic diversity on Earth. Despite the recent surge in their study, our knowledge on their actual biodiversity and distribution remains sparse. We report the first metagenomic analysis of Arctic freshwater viral DNA communities and a comparative analysis with other freshwater environments. Arctic viromes are dominated by unknown and single-stranded DNA viruses with no close relatives in the database. These unique viral DNA communities mostly relate to each other and present some minor genetic overlap with other environments studied, including an Arctic Ocean virome. Despite common environmental conditions in polar ecosystems, the Arctic and Antarctic DNA viromes differ at the fine-grain genetic level while sharing a similar taxonomic composition. The study uncovers some viral lineages with a bipolar distribution, suggesting a global dispersal capacity for viruses, and seemingly indicates that viruses do not follow the latitudinal diversity gradient known for macroorganisms. Our study sheds light into the global biogeography and connectivity of viral communities.
Keywords: Arctic; Virus; biodiversity; environmental microbiology; freshwater lake; metagenomics.
Figures



References
-
- Suttle C. A., Viruses in the sea. Nature 437, 356–361 (2005). - PubMed
-
- Ignacio-Espinoza J. C., Solonenko S. A., Sullivan M. B., The global virome: Not as big as we thought? Curr. Opin. Virol. 3, 566–571 (2013). - PubMed
-
- Fuhrman J. A., Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999). - PubMed
-
- Mann N. H., Cook A., Millard A., Bailey S., Clokie M., Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature 424, 741 (2003). - PubMed
-
- Williamson S. J., Rusch D. B., Yooseph S., Halpern A. L., Heidelberg K. B., Glass J. I., Andrews-Pfannkoch C., Fadrosh D., Miller C. S., Sutton G., Frazier M., Venter J. C., The Sorcerer II Global Ocean Sampling Expedition: Metagenomic characterization of viruses within aquatic microbial samples. PLOS One 3, e1456 (2008). - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

