A phylogenomic data-driven exploration of viral origins and evolution
- PMID: 26601271
- PMCID: PMC4643759
- DOI: 10.1126/sciadv.1500527
A phylogenomic data-driven exploration of viral origins and evolution
Abstract
The origin of viruses remains mysterious because of their diverse and patchy molecular and functional makeup. Although numerous hypotheses have attempted to explain viral origins, none is backed by substantive data. We take full advantage of the wealth of available protein structural and functional data to explore the evolution of the proteomic makeup of thousands of cells and viruses. Despite the extremely reduced nature of viral proteomes, we established an ancient origin of the "viral supergroup" and the existence of widespread episodes of horizontal transfer of genetic information. Viruses harboring different replicon types and infecting distantly related hosts shared many metabolic and informational protein structural domains of ancient origin that were also widespread in cellular proteomes. Phylogenomic analysis uncovered a universal tree of life and revealed that modern viruses reduced from multiple ancient cells that harbored segmented RNA genomes and coexisted with the ancestors of modern cells. The model for the origin and evolution of viruses and cells is backed by strong genomic and structural evidence and can be reconciled with existing models of viral evolution if one considers viruses to have originated from ancient cells and not from modern counterparts.
Keywords: fold; horizontal gene transfer; origin of life; phylogenetic analysis; protein domain; structure; taxonomy; tree of life; virus.
Figures

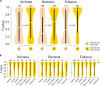
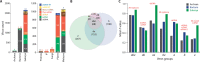




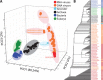
References
-
- Domingo E., Holland J. J., RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51, 151–178 (1997). - PubMed
-
- A. M. Q. King, M. J. Adams, E. B. Carstens, E. J. Lefkowitz, Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses (Elsevier, San Diego, CA, 2012).
-
- Krupovic M., Bamford D. H., Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr. Opin. Virol. 1, 118–124 (2011). - PubMed
-
- Balaji S., Srinivasan N., Comparison of sequence-based and structure-based phylogenetic trees of homologous proteins: Inferences on protein evolution. J. Biosci. 32, 83–96 (2007). - PubMed
-
- Abroi A., Gough J., Are viruses a source of new protein folds for organisms? Virosphere structure space and evolution. Bioessays 33, 626–635 (2011). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

